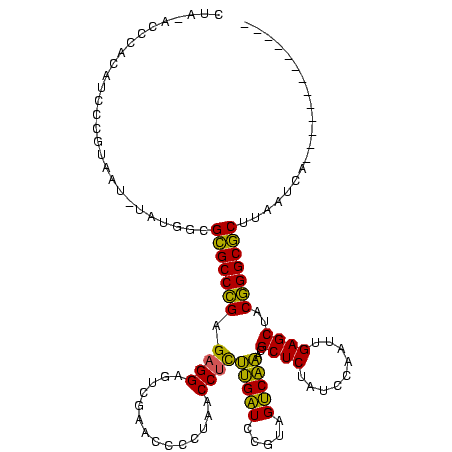
| Sequence ID | Bsubt.0 |
|---|---|
| Location | 634,589 – 634,709 |
| Length | 120 |
| Max. P | 0.999998 |

| Location | 634,589 – 634,695 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.53 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -29.54 |
| Energy contribution | -27.06 |
| Covariance contribution | -2.48 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.987003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 106 + 4214630/80-200 -------------CAAU-AAGCGCCCGUAGCUCAAUUGGAUAGAGCGUUUGACUACGGAUCAAAAGGUUAGGGGUUCGACUCCUCUCGGGCGCGCCAUAUAAGAUACCACGCGGGUGUAG -------------....-..(((((((..((((.........)))).((((((....).))))).....(((((.....)))))..)))))))..........(((((.....))))).. ( -34.00) >Blich.0 611619 106 + 4222334/80-200 -------------UGAUGAAGCGCCCGUAGCUCAAUUGGAUAGAGCGUUUGACUACGGAUCAAAAGGUUAGGGGUUCGACUCCUCUCGGGCGCGCCAUAUAUU-CGCGAUGCGGGUGUAG -------------.......(((((((..((((.........)))).((((((....).))))).....(((((.....)))))..)))))))....((((((-(((...))))))))). ( -37.50) >Banth.0 245805 116 + 5227293/80-200 CGAAAAAACAUCAUGAAUAAGCGCCCGUAGCUCAAUUGGAUAGAGCGUUUGACUACGGAUCAAGAGGUUAGGGGUUCGACUCCUCUCGGGCGCGCCA---AUUACGGGAAGUGGCU-CAG .............(((....((((((...((((.........))))..(((((....).))))(((...(((((.....))))))))))))))((((---.((.....)).)))))-)). ( -36.00) >Bcere.0 279198 115 + 5224283/80-200 CGAAAAA-CAUCAUGAAUAAGCGCCCGUAGCUCAAUUGGAUAGAGCGUUUGACUACGGAUCAAGAGGUUAGGGGUUCGACUCCUCUCGGGCGCGCCA---AUUACGGGAAGUGGCU-CAG .......-.....(((....((((((...((((.........))))..(((((....).))))(((...(((((.....))))))))))))))((((---.((.....)).)))))-)). ( -36.00) >Bhalo.0 558855 111 + 4202352/80-200 UUGCCUU------CAUUUAUGGGCCUGUAGCUCAGCUGGUUAGAGCGCACGCCUGAUAAGCGUGAGGUCGGUGGUUCAAGUCCACUCAGGCCCACCAUA-AAAAUAAAGUGUUGAC--AA .......------......((((((((...(((((((.(((((.((....))))))).))).))))...(((((.......))))))))))))).....-................--.. ( -36.50) >consensus _____________UGAUUAAGCGCCCGUAGCUCAAUUGGAUAGAGCGUUUGACUACGGAUCAAGAGGUUAGGGGUUCGACUCCUCUCGGGCGCGCCAUA_AUUACGCGAAGUGGCU_CAG ....................(((((((..((((.........)))).(((((.......))))).....(((((.......))))))))))))........................... (-29.54 = -27.06 + -2.48)

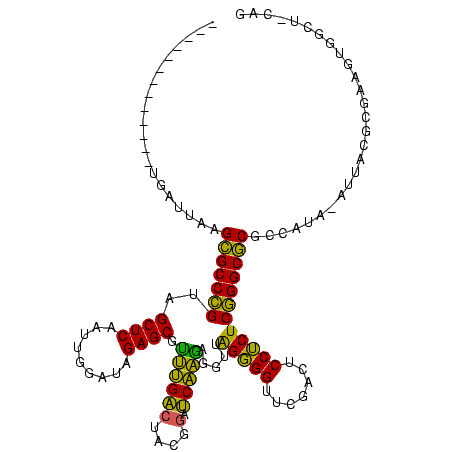
| Location | 634,589 – 634,695 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.53 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -27.04 |
| Energy contribution | -25.40 |
| Covariance contribution | -1.64 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 106 + 4214630/80-200 CUACACCCGCGUGGUAUCUUAUAUGGCGCGCCCGAGAGGAGUCGAACCCCUAACCUUUUGAUCCGUAGUCAAACGCUCUAUCCAAUUGAGCUACGGGCGCUU-AUUG------------- ........((((.((((...)))).))))(((((...(((.(((((..........))))))))..........((((.........))))..)))))....-....------------- ( -29.30) >Blich.0 611619 106 + 4222334/80-200 CUACACCCGCAUCGCG-AAUAUAUGGCGCGCCCGAGAGGAGUCGAACCCCUAACCUUUUGAUCCGUAGUCAAACGCUCUAUCCAAUUGAGCUACGGGCGCUUCAUCA------------- .......(((...)))-.....((((.(((((((...(((.(((((..........))))))))..........((((.........))))..))))))).))))..------------- ( -29.70) >Banth.0 245805 116 + 5227293/80-200 CUG-AGCCACUUCCCGUAAU---UGGCGCGCCCGAGAGGAGUCGAACCCCUAACCUCUUGAUCCGUAGUCAAACGCUCUAUCCAAUUGAGCUACGGGCGCUUAUUCAUGAUGUUUUUUCG .((-(((.....((((((.(---(((((((..(((((((..............)))))))...))).)))))..((((.........)))))))))).)))))................. ( -31.94) >Bcere.0 279198 115 + 5224283/80-200 CUG-AGCCACUUCCCGUAAU---UGGCGCGCCCGAGAGGAGUCGAACCCCUAACCUCUUGAUCCGUAGUCAAACGCUCUAUCCAAUUGAGCUACGGGCGCUUAUUCAUGAUG-UUUUUCG .((-(((.....((((((.(---(((((((..(((((((..............)))))))...))).)))))..((((.........)))))))))).))))).........-....... ( -31.94) >Bhalo.0 558855 111 + 4202352/80-200 UU--GUCAACACUUUAUUUU-UAUGGUGGGCCUGAGUGGACUUGAACCACCGACCUCACGCUUAUCAGGCGUGCGCUCUAACCAGCUGAGCUACAGGCCCAUAAAUG------AAGGCAA ..--.......((((((((.-....(((((((((.((((.......))))......((((((.....)))))).((((.........))))..))))))))))))))------))).... ( -39.10) >consensus CUA_ACCCACAUCCCGUAAU_UAUGGCGCGCCCGAGAGGAGUCGAACCCCUAACCUCUUGAUCCGUAGUCAAACGCUCUAUCCAAUUGAGCUACGGGCGCUUAAUCA_____________ ...........................(((((((.((((..............))))(((((.....)))))..((((.........))))..))))))).................... (-27.04 = -25.40 + -1.64)

| Location | 634,589 – 634,706 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.65 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -13.84 |
| Energy contribution | -13.60 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 117 + 4214630/360-480 GGGCCUGCACGAUGCAGGUCACACAGGUGUCACCGCGGGAUGCGGUGGACUUAACCUGUGAUCCAUUUAUC---GGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAA .(((((((.....))))))).(((((((.(((((((.....))))))).....))))))).(((.......---)))..(((((.(((((...))))))))))...(((.....)))... ( -51.20) >Blich.0 611619 120 + 4222334/360-480 GGGCCUGCACGAUGCAGGUCAUGCAGAUGUUGUCACAGGAUGUGACGAACUUAAUCUGCGCUCCAUUAUUUCAUGGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAA .(((((((.....)))))))..((((((.(((((((.....))))))).....)))))).((((((......)))))).(((((.(((((...))))))))))...(((.....)))... ( -50.90) >Banth.0 245805 115 + 5227293/360-480 AAAACUGAACGAAACAAACAACGUGAAA--CGUCAAUUUUUAUUUUAGAUGCUAGACAAACUAACUUUAUU---GGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAA ....................((((....--(((((.(.((((((((.((.(((((.((((((..((.....---))..))))))...))))))))))))))).).)))))))))...... ( -24.60) >Bcere.0 279198 115 + 5224283/360-480 AAAACUGAACGAAACAAACAACGUGAAA--CGUCAAUUUUUAUUUUAGAUGCUAGACAAACUAACUUUAUU---GGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAA ....................((((....--(((((.(.((((((((.((.(((((.((((((..((.....---))..))))))...))))))))))))))).).)))))))))...... ( -24.60) >Bhalo.0 558855 101 + 4202352/360-480 ---------UGAAGU--GUCACAGGAAA----CAACAUCCUGUG--AAACAUGAGCUUUAUCAAACUUUUA--UGGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAA ---------....((--.((((((((..----.....)))))))--).))..(((((..(((((((((((.--..)))))))))))...)))))(((....((((.....)))).))).. ( -39.10) >consensus AAAACUGAACGAAGCAAGUCACGCGAAA__CGUCACAUGAUGUGUUAGACGUAAGCUAAACUAAAUUUAUU___GGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAA ............................................................................((.(((((.(((((...)))))))))).))(((.....)))... (-13.84 = -13.60 + -0.24)

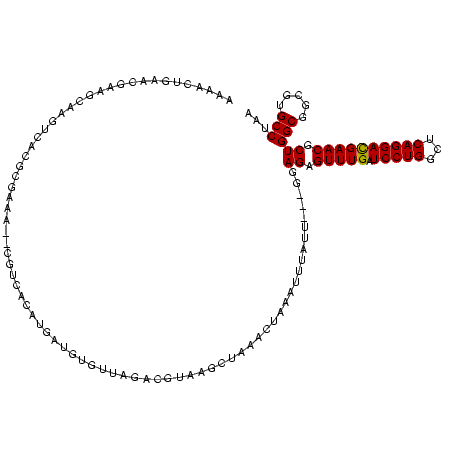
| Location | 634,589 – 634,707 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.03 |
| Mean single sequence MFE | -42.24 |
| Consensus MFE | -42.97 |
| Energy contribution | -41.20 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.77 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 118 + 4214630/480-600 UACAUGCAAGUCGAGCGAACAGAU-GGGAGCUUGCUCCC-UGAUGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAA ...((.(.((((..((.((((...-(((((....)))))-...)))).)).....((((((..((.((......)).))..))))))..)))).).))..((((((....)))))).... ( -45.60) >Blich.0 611619 118 + 4222334/480-600 UACAUGCAAGUCGAGCGGACCGAC-GGGAGCUUGCUCCC-UUAGGUCAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAA ...((.(.((((..((.((((...-(((((....)))))-...)))).)).....((((((..((.((......)).))..))))))..)))).).))..((((((....)))))).... ( -49.00) >Banth.0 245805 120 + 5227293/480-600 UACAUGCAAGUCGAGCGAAUGGAUUAAGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAA ...((.(.((((..((.(((....((((((....))))))....))).))(((((.((.(((.....))).))......))))).....)))).).))..((((((....)))))).... ( -38.90) >Bcere.0 279198 120 + 5224283/480-600 UACAUGCAAGUCGAGCGAAUGGAUUAAGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAA ...((.(.((((..((.(((....((((((....))))))....))).))(((((.((.(((.....))).))......))))).....)))).).))..((((((....)))))).... ( -38.90) >Bhalo.0 558855 118 + 4202352/480-600 UACAUGCAAGUCGAGCGGACCAAA-GGGAGCUUGCUUCU-GGAGGUUAGCGGCGAACGGGUGAGUAACACGUGGGCAACCUGCCUGUAAGACUGGGAUAACAUCGAGAAAUCGGUGCUAA ...((.(.((((..((.((((...-(((((....)))))-...)))).))((((.(((.((.....)).)))((....)))))).....)))).).))..((((((....)))))).... ( -38.80) >consensus UACAUGCAAGUCGAGCGAACCGAU_GGGAGCUUGCUCCU_UGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAA ...((.(.((((..((.((((...((((((....))))))...)))).))((((.(((.((.....)).)))((....)))))).....)))).).))..((((((....)))))).... (-42.97 = -41.20 + -1.77)

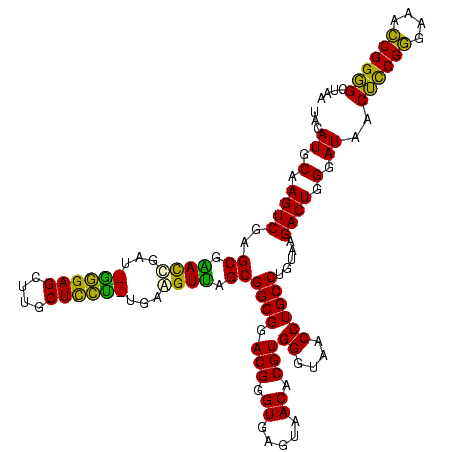
| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.25 |
| Mean single sequence MFE | -41.32 |
| Consensus MFE | -41.68 |
| Energy contribution | -38.00 |
| Covariance contribution | -3.68 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.61 |
| Structure conservation index | 1.01 |
| SVM decision value | 4.22 |
| SVM RNA-class probability | 0.999840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/520-640 UGAUGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGCAUGGUUCAAACAUAAAA .(.((((((((((..((((((..((.((......)).))..))))))..).)))......((((((....)))))))))))).).......((((((((((....))))))))))..... ( -43.80) >Blich.0 611619 120 + 4222334/520-640 UUAGGUCAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAGCCGCAUGGUUCAAUUAUAAAA .......((((.(..((((((..((.((......)).))..))))))..).((((..((.((((((....)))))).))...)))).))))((((((((((....))))))))))..... ( -44.00) >Banth.0 245805 120 + 5227293/520-640 UGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAA ....((((.(((....((((((.....)))((((((.....))))))....)))......((((((....))))))......)))..))))((((((((((....))))))))))..... ( -41.80) >Bcere.0 279198 120 + 5224283/520-640 UGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAA ....((((.(((....((((((.....)))((((((.....))))))....)))......((((((....))))))......)))..))))((((((((((....))))))))))..... ( -41.80) >Bhalo.0 558855 120 + 4202352/520-640 GGAGGUUAGCGGCGAACGGGUGAGUAACACGUGGGCAACCUGCCUGUAAGACUGGGAUAACAUCGAGAAAUCGGUGCUAAUACCGGAUAAUAAAAAGAACUGCAUGGUUCUUUUUUGAAA ..((((..(..((..(((.((.....)).)))..))..)..))))......((((..((.((((((....)))))).))...)))).....((((((((((....))))))))))..... ( -35.20) >consensus UGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAAUAGUUUGAACCGCAUGGUUCAAAAUUGAAA ....((((..((((.(((.((.....)).)))((....)))))).......((((..((.((((((....)))))).))...)))).))))((((((((((....))))))))))..... (-41.68 = -38.00 + -3.68)

| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.25 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -27.56 |
| Energy contribution | -26.60 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/520-640 UUUUAUGUUUGAACCAUGCGGUUCAAACAACCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUAACAUCA .....((((((((((....)))))))))).......(((.(((((.((((....))))..................(((.((....))..(((.....)))......)))))))).))). ( -34.60) >Blich.0 611619 120 + 4222334/520-640 UUUUAUAAUUGAACCAUGCGGCUCAAUCAAGCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUGACCUAA ..............((.(((((..............((((.((((.((((....)))).))))..((.(((.....))).)).))))...(((.....)))......)))))))...... ( -28.10) >Banth.0 245805 120 + 5227293/520-640 UUUCAAUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUAACUUCA .....((((.(((((....))))).))))((((..((((..((((.((((....)))).))))............((((.((....))..(((.....)))..)))))))).)))).... ( -31.10) >Bcere.0 279198 120 + 5224283/520-640 UUUCAAUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUAACUUCA .....((((.(((((....))))).))))((((..((((..((((.((((....)))).))))............((((.((....))..(((.....)))..)))))))).)))).... ( -31.10) >Bhalo.0 558855 120 + 4202352/520-640 UUUCAAAAAAGAACCAUGCAGUUCUUUUUAUUAUCCGGUAUUAGCACCGAUUUCUCGAUGUUAUCCCAGUCUUACAGGCAGGUUGCCCACGUGUUACUCACCCGUUCGCCGCUAACCUCC .....(((((((((......))))))))).......((...(((((.(((....))).)))))..)).........((.(((((((....(((..((......)).))).)).))))))) ( -24.30) >consensus UUUCAAAUUCGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUAACCUCA .....((((((((((....)))))))))).......((...((((.((((....)))).))))..)).........(((.((....))..(((.....)))......))).......... (-27.56 = -26.60 + -0.96)

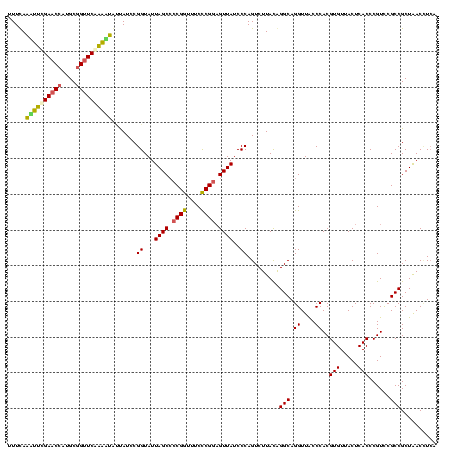
| Location | 634,589 – 634,708 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.84 |
| Mean single sequence MFE | -49.54 |
| Consensus MFE | -51.68 |
| Energy contribution | -46.56 |
| Covariance contribution | -5.12 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.97 |
| Structure conservation index | 1.04 |
| SVM decision value | 4.55 |
| SVM RNA-class probability | 0.999919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 119 + 4214630/560-680 UGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGCAUGGUUCAAACAUAAAAGGUGGCUU-CGGCUACCACUUACAGAUGGACCCGCGGCGC .(((((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((..-..)))))).))))))(.((....)))))).. ( -55.20) >Blich.0 611619 120 + 4222334/560-680 UGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAGCCGCAUGGUUCAAUUAUAAAAGGUGGCUUUCAGCUACCACUUACAGAUGGACCCGCGGCGC .(((((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((.....)))))).))))))(.((....)))))).. ( -50.30) >Banth.0 245805 119 + 5227293/560-680 UGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUU-CGGCUGUCACUUAUGGAUGGACCCGCGUCGC ...(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((..-..)))))).)))))))...(((....))).. ( -51.30) >Bcere.0 279198 119 + 5224283/560-680 UGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUU-CGGCUGUCACUUAUGGAUGGACCCGCGUCGC ...(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((..-..)))))).)))))))...(((....))).. ( -51.30) >Bhalo.0 558855 119 + 4202352/560-680 UGCCUGUAAGACUGGGAUAACAUCGAGAAAUCGGUGCUAAUACCGGAUAAUAAAAAGAACUGCAUGGUUCUUUUUUGAAAGAUGGUUU-CGGCUAUCACUUACAGAUGGGCCCGCGGCGC ...(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((..-..)))))).)))))))....(((...))).. ( -39.60) >consensus UGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAAUAGUUUGAACCGCAUGGUUCAAAAUUGAAAGGUGGCUU_CGGCUAUCACUUACAGAUGGACCCGCGGCGC .(((((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((.....)))))).)))))))..(....)))..... (-51.68 = -46.56 + -5.12)

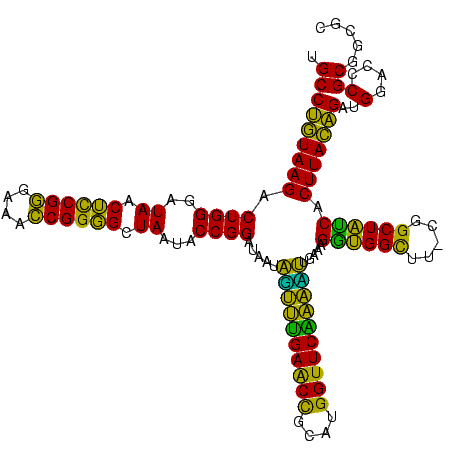
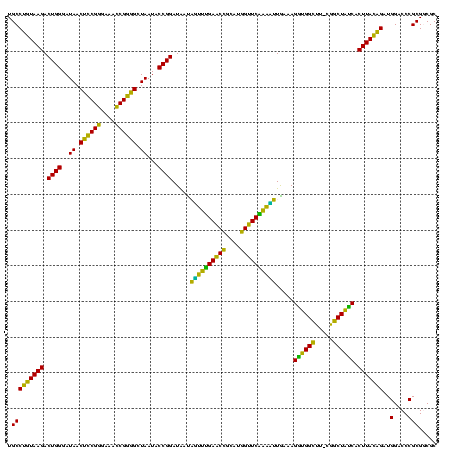
| Location | 634,589 – 634,708 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.68 |
| Mean single sequence MFE | -42.46 |
| Consensus MFE | -41.02 |
| Energy contribution | -36.90 |
| Covariance contribution | -4.12 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 119 + 4214630/600-720 UACCGGAUGGUUGUUUGAACCGCAUGGUUCAAACAUAAAAGGUGGCUU-CGGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCAACGA ..(((..((..((((((((((....)))))))))).....((((((..-..)))))).....))..)))...((..((((....))...((((((((.......)))))))).))..)). ( -48.10) >Blich.0 611619 120 + 4222334/600-720 UACCGGAUGCUUGAUUGAGCCGCAUGGUUCAAUUAUAAAAGGUGGCUUUCAGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGA ...(((((((((....))(((((..((((((.((.(((..((((((.....))))))..))).)).)))))).)))))))))).(((..((((((((.......)))))))))))..)). ( -45.60) >Banth.0 245805 119 + 5227293/600-720 UACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUU-CGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGA ..(((......((((((((((....)))))))))).....((((((..-..)))))).....)))((((((....))).)))..(((..((((((((.......)))))))))))..... ( -40.90) >Bcere.0 279198 119 + 5224283/600-720 UACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUU-CGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGA ..(((......((((((((((....)))))))))).....((((((..-..)))))).....)))((((((....))).)))..(((..((((((((.......)))))))))))..... ( -40.90) >Bhalo.0 558855 119 + 4202352/600-720 UACCGGAUAAUAAAAAGAACUGCAUGGUUCUUUUUUGAAAGAUGGUUU-CGGCUAUCACUUACAGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGA .((((......((((((((((....)))))))))).......)))).(-((.((...(((..(.((((.(((...))).)))).)..)))(((((((.......))))))))).)))... ( -36.82) >consensus UACCGGAUAAUAGUUUGAACCGCAUGGUUCAAAAUUGAAAGGUGGCUU_CGGCUAUCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGA ..(((..((..((((((((((....)))))))))).....((((((.....)))))).....))..)))....((...((....))...((((((((.......)))))))).))..... (-41.02 = -36.90 + -4.12)

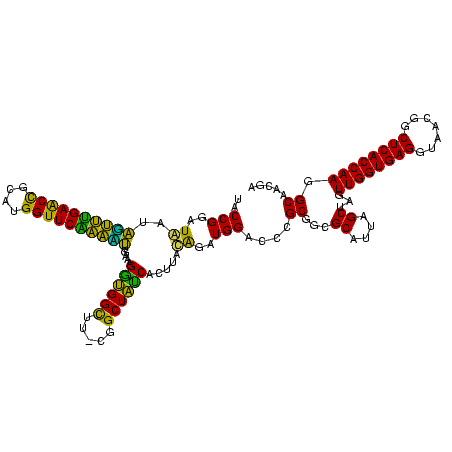
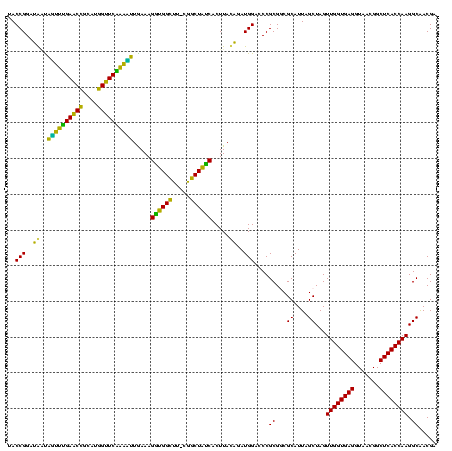
| Location | 634,589 – 634,708 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.68 |
| Mean single sequence MFE | -35.73 |
| Consensus MFE | -28.80 |
| Energy contribution | -27.88 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 119 + 4214630/600-720 UCGUUGCCUUGGUGAGCCAUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGGUAGCCG-AAGCCACCUUUUAUGUUUGAACCAUGCGGUUCAAACAACCAUCCGGUA .((((((.((((((((.......))))))))...)).)))).((((..(((..........(((((.....-..)))))......((((((((((....)))))))))))))...)))). ( -39.90) >Blich.0 611619 120 + 4222334/600-720 UCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGGUAGCUGAAAGCCACCUUUUAUAAUUGAACCAUGCGGCUCAAUCAAGCAUCCGGUA .....(((((((((((.......))))))))...(((..((.(((((((((.((..((((((.(((.((.....)).))).))))))..)).))).)))))).))....)))....))). ( -43.50) >Banth.0 245805 119 + 5227293/600-720 UCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCCG-AAGCCGCCUUUCAAUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUA .((((((.((((((((.......)))))))).........))))))(((((((........).)))..)))-..((((.......((((.(((((....))))).))))......)))). ( -31.42) >Bcere.0 279198 119 + 5224283/600-720 UCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCCG-AAGCCGCCUUUCAAUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUA .((((((.((((((((.......)))))))).........))))))(((((((........).)))..)))-..((((.......((((.(((((....))))).))))......)))). ( -31.42) >Bhalo.0 558855 119 + 4202352/600-720 UCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGCCCAUCUGUAAGUGAUAGCCG-AAACCAUCUUUCAAAAAAGAACCAUGCAGUUCUUUUUAUUAUCCGGUA ..(((((.((((((((.......))))))))...((..(((.(((...))).)))..))..))))).((((-(((.....)))).(((((((((......))))))))).......))). ( -32.40) >consensus UCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGAUAGCCG_AAGCCACCUUUCAAAUUCGAACCAUGCGGUUCAAAAUAUUAUCCGGUA .(((.((.((((((((.......))))))))...((....)))).)))(((.(((......)))...)))....(((........((((((((((....)))))))))).......))). (-28.80 = -27.88 + -0.92)

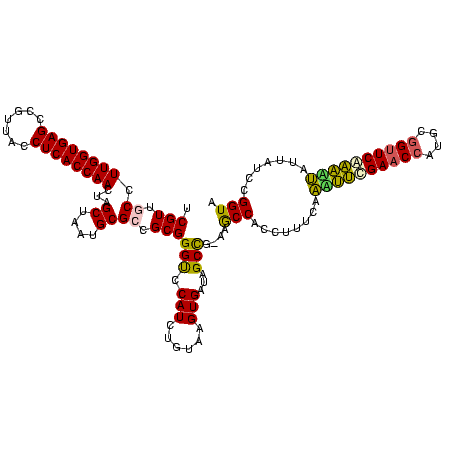
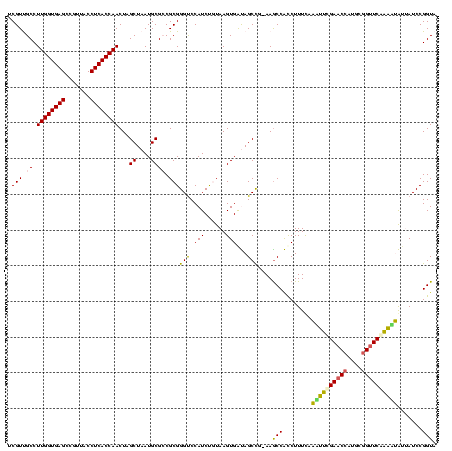
| Location | 634,589 – 634,708 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -51.72 |
| Consensus MFE | -45.64 |
| Energy contribution | -47.10 |
| Covariance contribution | 1.46 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 119 + 4214630/640-760 GGUGGCUU-CGGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((..-..)))))).............(((...(((((((.(((..((((((((.......)))))))))))...))))))).((((((((....)))...)))))......))).. ( -52.40) >Blich.0 611619 120 + 4222334/640-760 GGUGGCUUUCAGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((.....)))))).............(((...(((((((.(((..((((((((.......)))))))))))...))))))).((((((((....)))...)))))......))).. ( -50.40) >Banth.0 245805 119 + 5227293/640-760 GGCGGCUU-CGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((..-..)))))).............(((((((((.....(((..((((((((.......)))))))))))..))))))((.((((((((....)))...))))).))...))).. ( -52.70) >Bcere.0 279198 119 + 5224283/640-760 GGCGGCUU-CGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((..-..)))))).............(((((((((.....(((..((((((((.......)))))))))))..))))))((.((((((((....)))...))))).))...))).. ( -52.70) >Bhalo.0 558855 119 + 4202352/640-760 GAUGGUUU-CGGCUAUCACUUACAGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((..-..)))))).....(((.((((((.((.(((((((.(((..((((((((.......)))))))))))...))))))).))..((((....))))....)))).))))).... ( -50.40) >consensus GGUGGCUU_CGGCUAUCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((.....)))))).............(((.....(((((.(((..((((((((.......)))))))))))...)))))((.((((((((....)))...))))).))...))).. (-45.64 = -47.10 + 1.46)

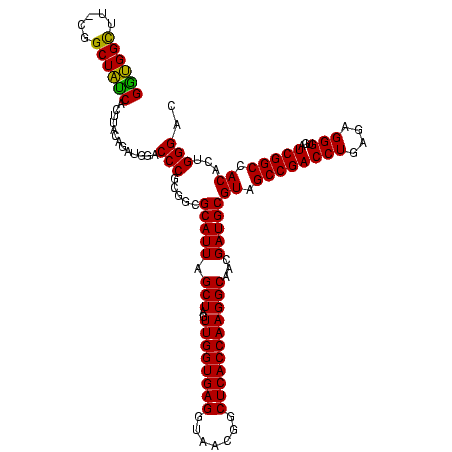
| Location | 634,589 – 634,708 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -43.96 |
| Consensus MFE | -39.82 |
| Energy contribution | -39.42 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 119 + 4214630/640-760 GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCAUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGGUAGCCG-AAGCCACC (.(((.((((((((((((.........))))))))))))...((.((.((((((((.......))))))))...((....)))).))))).).........(((((.....-..))))). ( -46.80) >Blich.0 611619 120 + 4222334/640-760 GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGGUAGCUGAAAGCCACC (.(((.((((((((((((.........))))))))))))...((.((.((((((((.......))))))))...((....)))).))))).).........(((((........))))). ( -47.10) >Banth.0 245805 119 + 5227293/640-760 GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCCG-AAGCCGCC (.(((.((((((((((((.........)))))))))))).((((.((.((((((((.......))))))))...))....))))...))).).........(((...((..-..))))). ( -41.50) >Bcere.0 279198 119 + 5224283/640-760 GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCCG-AAGCCGCC (.(((.((((((((((((.........)))))))))))).((((.((.((((((((.......))))))))...))....))))...))).).........(((...((..-..))))). ( -41.50) >Bhalo.0 558855 119 + 4202352/640-760 GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGCCCAUCUGUAAGUGAUAGCCG-AAACCAUC ......((((((((((((.........))))))))))))..(((.((.((((((((.......))))))))...((....)))).)))(((..(((.......))).))).-........ ( -42.90) >consensus GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGAUAGCCG_AAGCCACC (.(((.((((((((((((.........))))))))))))......((.((((((((.......))))))))...((....))...))))).).........(((............))). (-39.82 = -39.42 + -0.40)

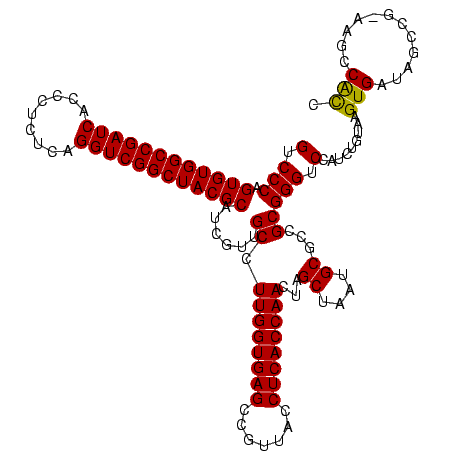
| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -51.64 |
| Consensus MFE | -46.64 |
| Energy contribution | -51.64 |
| Covariance contribution | 5.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/680-800 AUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... ( -51.60) >Blich.0 611619 120 + 4222334/680-800 AUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... ( -51.70) >Banth.0 245805 120 + 5227293/680-800 AUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... ( -51.60) >Bcere.0 279198 120 + 5224283/680-800 AUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... ( -51.60) >Bhalo.0 558855 120 + 4202352/680-800 AUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... ( -51.70) >consensus AUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... (-46.64 = -51.64 + 5.00)


| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -43.36 |
| Consensus MFE | -43.12 |
| Energy contribution | -43.12 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/680-800 CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCAUUACCUCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))...((((..((((((((.......)))))))).)))).... ( -43.40) >Blich.0 611619 120 + 4222334/680-800 CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))......((.((((((((.......))))))))...)).... ( -43.30) >Banth.0 245805 120 + 5227293/680-800 CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))...((((..((((((((.......)))))))).)))).... ( -43.40) >Bcere.0 279198 120 + 5224283/680-800 CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))...((((..((((((((.......)))))))).)))).... ( -43.40) >Bhalo.0 558855 120 + 4202352/680-800 CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))......((.((((((((.......))))))))...)).... ( -43.30) >consensus CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))......((.((((((((.......))))))))...)).... (-43.12 = -43.12 + 0.00)

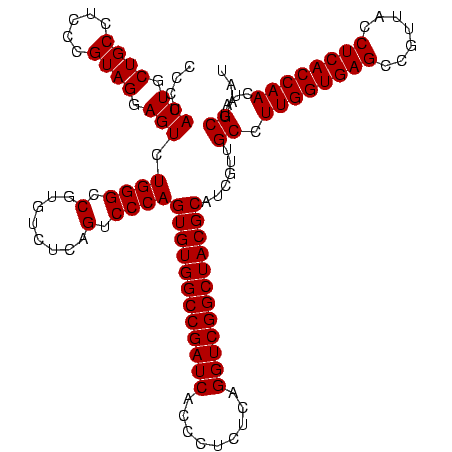
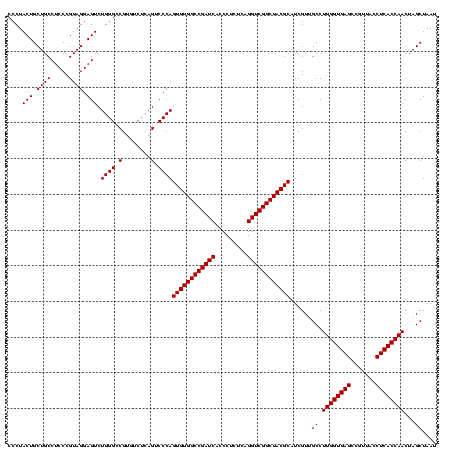
| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -49.80 |
| Consensus MFE | -44.80 |
| Energy contribution | -49.80 |
| Covariance contribution | 5.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/720-840 UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). ( -49.80) >Blich.0 611619 120 + 4222334/720-840 UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). ( -49.80) >Banth.0 245805 120 + 5227293/720-840 UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). ( -49.80) >Bcere.0 279198 120 + 5224283/720-840 UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). ( -49.80) >Bhalo.0 558855 120 + 4202352/720-840 UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). ( -49.80) >consensus UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). (-44.80 = -49.80 + 5.00)

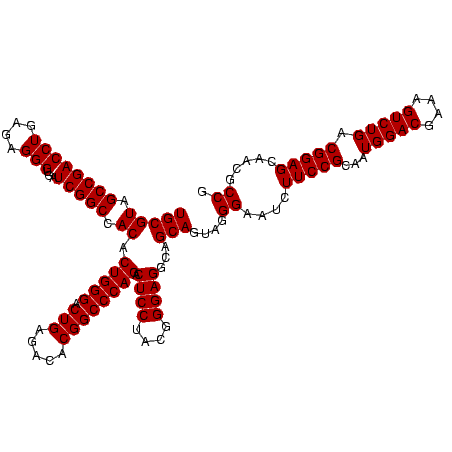
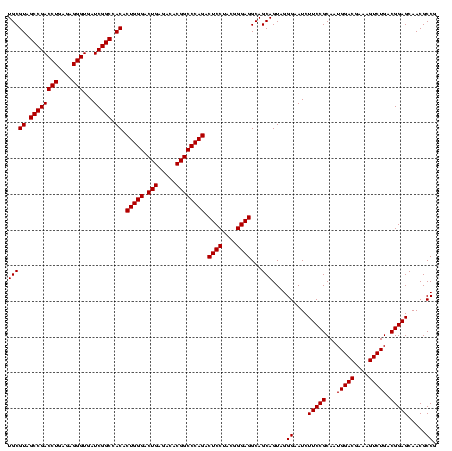
| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -46.94 |
| Consensus MFE | -44.96 |
| Energy contribution | -45.32 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.84 |
| Structure conservation index | 0.96 |
| SVM decision value | 5.64 |
| SVM RNA-class probability | 0.999991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/800-920 AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUG ..((((((.....((((....))))((((((((..((.(((....))).))..((((....)))).....)))))))).....)))))).((((((((((((.....)))))))).)))) ( -48.90) >Blich.0 611619 120 + 4222334/800-920 AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUG ..((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....)))))).((((((((((((.....)))))))).)))) ( -44.50) >Banth.0 245805 120 + 5227293/800-920 AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUG ..((((((.....((((....))))(((((((..(((((.((.((((.......)))).)))).)))....))))))).....)))))).((((((((((((.....)))))))).)))) ( -45.90) >Bcere.0 279198 120 + 5224283/800-920 AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUG ..((((((.....((((....))))(((((((..(((((.((.((((.......)))).)))).)))....))))))).....)))))).((((((((((((.....)))))))).)))) ( -45.90) >Bhalo.0 558855 119 + 4202352/800-920 AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAA-AGGGCGGCACCUUG ..((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....)))))).((((((((((((...-.)))))))).)))) ( -49.50) >consensus AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAAUAGGGCGGCACCUUG ..((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....)))))).((((((((((((.....)))))))).)))) (-44.96 = -45.32 + 0.36)

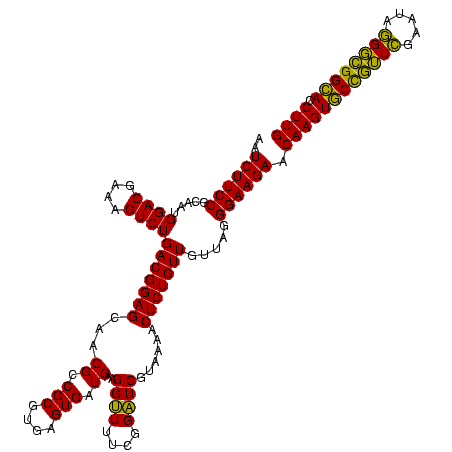
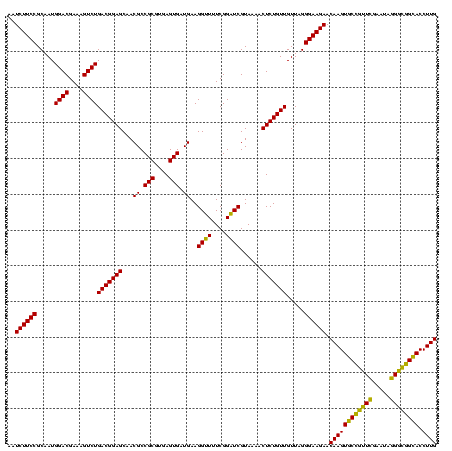
| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -40.26 |
| Consensus MFE | -39.34 |
| Energy contribution | -36.70 |
| Covariance contribution | -2.64 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.68 |
| SVM RNA-class probability | 0.999517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/840-960 CGUGAGUGAUGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAG .(((.((......((((...(((((((....(((((....))))).....((((((((((((.....)))))))).))))))))).)).)))).....)))))(((.........))).. ( -37.90) >Blich.0 611619 120 + 4222334/840-960 CGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAG .(((.((......((((...(((((((....(((((....))))).....((((((((((((.....)))))))).))))))))).)).)))).....)))))(((.........))).. ( -38.20) >Banth.0 245805 120 + 5227293/840-960 CGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAG (....).......(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..)))))))..(((.........))).. ( -40.90) >Bcere.0 279198 120 + 5224283/840-960 CGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAG (....).......(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..)))))))..(((.........))).. ( -40.90) >Bhalo.0 558855 119 + 4202352/840-960 CGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAA-AGGGCGGCACCUUGACGGUACCUAACGAGAAAGCCACGGCUAACUACGUGCCAG (....).......((((((((((.........))).((((((((......((((((((((((...-.)))))))).))))......)))))))))))))))..(((.........))).. ( -43.40) >consensus CGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAAUAGGGCGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAG .............((((((((((.........)))...((((((......((((((((((((.....)))))))).))))......))))))..)))))))..(((.........))).. (-39.34 = -36.70 + -2.64)

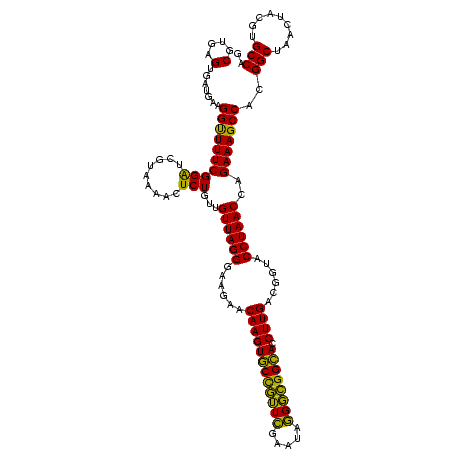
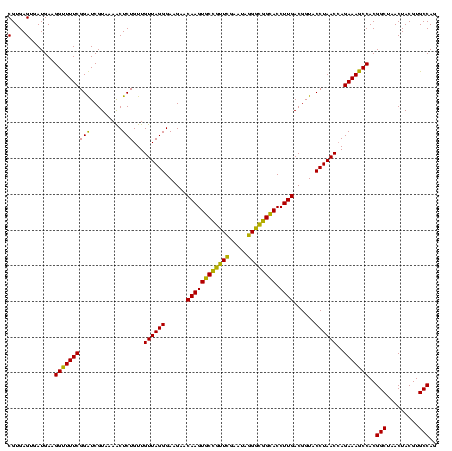
| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -41.96 |
| Consensus MFE | -42.68 |
| Energy contribution | -40.72 |
| Covariance contribution | -1.96 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.00 |
| Structure conservation index | 1.02 |
| SVM decision value | 4.01 |
| SVM RNA-class probability | 0.999754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/880-1000 UAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAU ((((......((((((((((((.....)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... ( -41.40) >Blich.0 611619 120 + 4222334/880-1000 UAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAU ((((......((((((((((((.....)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... ( -41.40) >Banth.0 245805 120 + 5227293/880-1000 UAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUAUCCGGAAU ((((......((((((((((((.....)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... ( -40.70) >Bcere.0 279198 120 + 5224283/880-1000 UAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUAUCCGGAAU ((((......((((((((((((.....)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... ( -40.70) >Bhalo.0 558855 119 + 4202352/880-1000 UAGGGAAGAACAAGUGCCGUUCGAA-AGGGCGGCACCUUGACGGUACCUAACGAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAU ((((......((((((((((((...-.)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)))).... ( -45.60) >consensus UAGGGAAGAACAAGUGCCGUUCGAAUAGGGCGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAU ((((......((((((((((((.....)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... (-42.68 = -40.72 + -1.96)

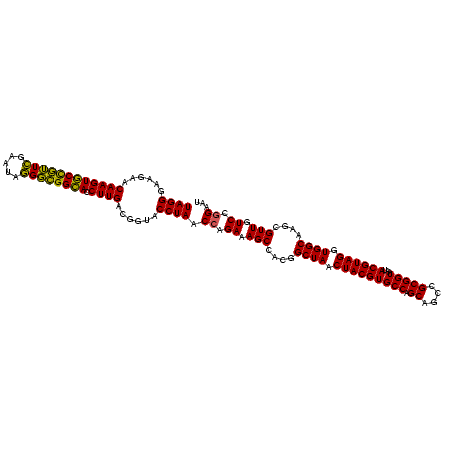
| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -38.42 |
| Consensus MFE | -36.52 |
| Energy contribution | -36.44 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/880-1000 AUUCCGGACAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGUACUUGUUCUUCCCUA .....(((.((((..((((............((((((((((((.....))).)))))))))......(((..((((((.....)))))))))..........))))..))))..)))... ( -38.60) >Blich.0 611619 120 + 4222334/880-1000 AUUCCGGACAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGUACUUGUUCUUCCCUA .....(((.((((..((((............((((((((((((.....))).)))))))))......(((..((((((.....)))))))))..........))))..))))..)))... ( -38.60) >Banth.0 245805 120 + 5227293/880-1000 AUUCCGGAUAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCAGCUUAUUCAACUAGCACUUGUUCUUCCCUA .....(((.((((..(((.............((((((((((((.....))).)))))))))......((((.((((((.....))))))))))..........)))..))))..)))... ( -37.20) >Bcere.0 279198 120 + 5224283/880-1000 AUUCCGGAUAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCAGCUUAUUCAACUAGCACUUGUUCUUCCCUA .....(((.((((..(((.............((((((((((((.....))).)))))))))......((((.((((((.....))))))))))..........)))..))))..)))... ( -37.20) >Bhalo.0 558855 119 + 4202352/880-1000 AUUCCGGACAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUCGUUAGGUACCGUCAAGGUGCCGCCCU-UUCGAACGGCACUUGUUCUUCCCUA .....((((((..(((((.(((((((.....((((((((((((.....))).)))))))))......)).)))))...).))))((((((..(.-...)..))))))))))))....... ( -40.50) >consensus AUUCCGGACAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCGCCCUAUUCGAACGGCACUUGUUCUUCCCUA .....(((.((((..((((............((((((((((((.....))).)))))))))...........((((((.....)))))).............))))..))))..)))... (-36.52 = -36.44 + -0.08)


| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -37.26 |
| Energy contribution | -36.14 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/1000-1120 UAUUGGGCGUAAAGGGCUCGCAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGAAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCA ....((((.......))))(((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))......((((.......)))). ( -38.30) >Blich.0 611619 120 + 4222334/1000-1120 UAUUGGGCGUAAAGCGCGCGCAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCA ....(.(((.....))).)(((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))......((((.......)))). ( -39.20) >Banth.0 245805 120 + 5227293/1000-1120 UAUUGGGCGUAAAGCGCGCGCAGGUGGUUUCUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCA ......(((.....)))((((....(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))...))))......((((.......)))). ( -38.10) >Bcere.0 279198 120 + 5224283/1000-1120 UAUUGGGCGUAAAGCGCGCGCAGGUGGUUUCUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCA ......(((.....)))((((....(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))...))))......((((.......)))). ( -38.10) >Bhalo.0 558855 120 + 4202352/1000-1120 UAUUGGGCGUAAAGCGCGCGCAGGCGGUCUCUUAAGUCUGAUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGAGACUUGAGUACAGAAGAGGAGAGUGGAAUUCCA ..(((.((((.....)))).))).((.((((((...((((..((((....((((((.....))))))....)))).....(((((....))..))).))))..)))))).))........ ( -40.80) >consensus UAUUGGGCGUAAAGCGCGCGCAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCA ....(.(((.....))).)(((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))......((((.......)))). (-37.26 = -36.14 + -1.12)

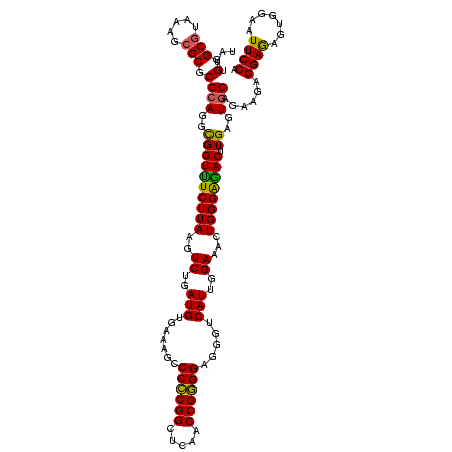
| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -35.08 |
| Energy contribution | -33.80 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.52 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/1040-1160 AUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGAAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCA ........((((((((.....))))))((..((...(....)...))..)).....)).........(.(((...((((((((.(.(((......)))....).))))))))...)))). ( -36.70) >Blich.0 611619 120 + 4222334/1040-1160 AUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCA ........((((((((.....))))))((..((...(....)...))..)).....)).........(.(((...((((((((.(.(((......)))....).))))))))...)))). ( -35.80) >Banth.0 245805 120 + 5227293/1040-1160 AUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCA ........((((((((.....)))))).(((((...(....).....)))))....))...........(((...((((((((.(.(((......)))....).))))))))...))).. ( -32.20) >Bcere.0 279198 120 + 5224283/1040-1160 AUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCA ........((((((((.....)))))).(((((...(....).....)))))....))...........(((...((((((((.(.(((......)))....).))))))))...))).. ( -32.20) >Bhalo.0 558855 120 + 4202352/1040-1160 AUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGAGACUUGAGUACAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCA .((((.....((((((.....)))))).(((((...(....).....)))))...))))........(.(((...((((((((.(.(((......)))....).))))))))...)))). ( -36.70) >consensus AUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCA .((((.....((((((.....))))))((..((...(....)...))..))....))))..........(((...((((((((.(.(((......)))....).))))))))...))).. (-35.08 = -33.80 + -1.28)

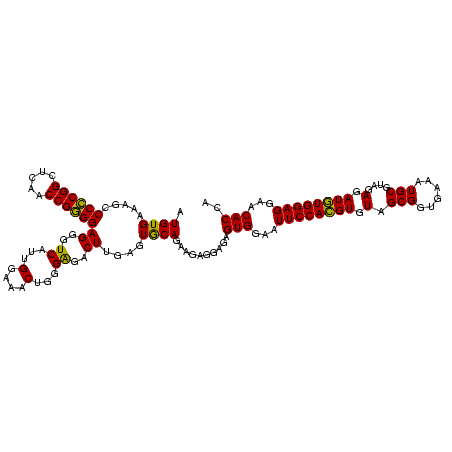
| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -38.96 |
| Consensus MFE | -39.78 |
| Energy contribution | -38.66 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.48 |
| Structure conservation index | 1.02 |
| SVM decision value | 4.88 |
| SVM RNA-class probability | 0.999958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/1080-1200 ACUGGGAAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGG .((.((.....(.((((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).)..)).)). ( -41.00) >Blich.0 611619 120 + 4222334/1080-1200 ACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGG .((.((...(.(.((((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).).))).)). ( -40.00) >Banth.0 245805 120 + 5227293/1080-1200 ACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGG .(.((....)).)((((((((..(((((.(((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).......... ( -36.40) >Bcere.0 279198 120 + 5224283/1080-1200 ACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGG .(.((....)).)((((((((..(((((.(((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).......... ( -36.40) >Bhalo.0 558855 120 + 4202352/1080-1200 ACUGGGAGACUUGAGUACAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGG .((.((.....(.((((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).)..)).)). ( -41.00) >consensus ACUGGGAGACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGG .((.((.....(.((((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).)..)).)). (-39.78 = -38.66 + -1.12)


| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -40.44 |
| Consensus MFE | -40.20 |
| Energy contribution | -40.00 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/1160-1280 GUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGAGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGU ((.((....)).))..............((((((((....((((...((((((...((...((((.........))))...))))))))((....)).....)))).))))))))..... ( -37.10) >Blich.0 611619 120 + 4222334/1160-1280 GUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGU ...((....)).((((((((((.....)))((((((..(((((....((((((...((...((((.........))))...))))))))((....))....))))).))))))))))))) ( -41.20) >Banth.0 245805 120 + 5227293/1160-1280 GUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGU ...((....))((((....)))).....((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))..... ( -41.60) >Bcere.0 279198 120 + 5224283/1160-1280 GUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGU ...((....))((((....)))).....((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))..... ( -41.60) >Bhalo.0 558855 120 + 4202352/1160-1280 GUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUU ((.((....)).))..............((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))..... ( -40.70) >consensus GUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGU ...((....))((((....)))).....((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))..... (-40.20 = -40.00 + -0.20)

| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -44.60 |
| Consensus MFE | -45.44 |
| Energy contribution | -44.32 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.47 |
| Structure conservation index | 1.02 |
| SVM decision value | 5.27 |
| SVM RNA-class probability | 0.999981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/1200-1320 AGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCC .(((...((((((...((...((((.........))))...))))))))))).......(((((((.((..(((((((((....)))))))))..))...((....))....))))))). ( -48.90) >Blich.0 611619 120 + 4222334/1200-1320 CGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGCAGCAAACGCAUUAAGCACUCC .(((...((((((...((...((((.........))))...))))))))))).......(((((((.((..(((((((((....)))))))))..))...((....))....))))))). ( -45.00) >Banth.0 245805 120 + 5227293/1200-1320 CGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCC .(((...(((((((..((...((((.........))))))..)))))))))).......(((((((.((..(((((((((....)))))))))..))...((....))....))))))). ( -42.40) >Bcere.0 279198 120 + 5224283/1200-1320 CGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCC .(((...(((((((..((...((((.........))))))..)))))))))).......(((((((.((..(((((((((....)))))))))..))...((....))....))))))). ( -42.40) >Bhalo.0 558855 120 + 4202352/1200-1320 CGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGAUGCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCC .(((...(((((((..((...((((.........))))))..)))))))))).......(((((((.((..((((((((......))))))))..))...((....))....))))))). ( -44.30) >consensus CGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCC .(((...(((((((..((...((((.........))))))..)))))))))).......(((((((.((..((((((((......))))))))..))...((....))....))))))). (-45.44 = -44.32 + -1.12)

| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -37.88 |
| Consensus MFE | -34.58 |
| Energy contribution | -33.82 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/1200-1320 GGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUCGCUCCCCACGCUUUCGCU .(((((((....(.((((((....)).....((((.(....))))))))).)..))))))).........(((((((.....((((((.....))))))........)))))))...... ( -42.72) >Blich.0 611619 120 + 4222334/1200-1320 GGAGUGCUUAAUGCGUUUGCUGCAGCACUAAAGGGCGGAAACCCUCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUCGCUCCCCACGCUUUCGCG .(((((((....(.((((((....)))....((((.(....).))))))).)..)))))))..(((....(((((((.....((((((.....))))))........)))))))...))) ( -37.12) >Banth.0 245805 120 + 5227293/1200-1320 GGAGUGCUUAAUGCGUUAACUUCAGCACUAAAGGGCGGAAACCCUCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCG .(((((((((.(((..........))).)))((((.(....).))))........))))))..(((....(((((((.....((((((.....))))))........)))))))...))) ( -35.72) >Bcere.0 279198 120 + 5224283/1200-1320 GGAGUGCUUAAUGCGUUAACUUCAGCACUAAAGGGCGGAAACCCUCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCG .(((((((((.(((..........))).)))((((.(....).))))........))))))..(((....(((((((.....((((((.....))))))........)))))))...))) ( -35.72) >Bhalo.0 558855 120 + 4202352/1200-1320 GGAGUGCUUAAUGUGUUAACUUCGGCACUAAGGGCAUCGAAACCCCUAACACCUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCG .(((((((....((((((.(....)......(((........))).))))))..)))))))..(((....(((((((.....((((((.....))))))........)))))))...))) ( -38.12) >consensus GGAGUGCUUAAUGCGUUAACUUCAGCACUAAAGGGCGGAAACCCUCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCG .(((((((((.(((..........))).)))((((.(....).))))........))))))...((....(((((((.....((((((.....))))))........)))))))...)). (-34.58 = -33.82 + -0.76)

| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -47.64 |
| Consensus MFE | -49.40 |
| Energy contribution | -47.48 |
| Covariance contribution | -1.92 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.49 |
| Structure conservation index | 1.04 |
| SVM decision value | 6.41 |
| SVM RNA-class probability | 0.999998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/1240-1360 AGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ ( -50.90) >Blich.0 611619 120 + 4222334/1240-1360 AGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGCAGCAAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ ( -47.00) >Banth.0 245805 120 + 5227293/1240-1360 AGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ ( -46.80) >Bcere.0 279198 120 + 5224283/1240-1360 AGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ ( -46.80) >Bhalo.0 558855 120 + 4202352/1240-1360 AGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGAUGCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..((((((((......))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ ( -46.70) >consensus AGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..((((((((......))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ (-49.40 = -47.48 + -1.92)

| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -43.68 |
| Consensus MFE | -40.90 |
| Energy contribution | -40.46 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/1280-1400 UUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAU ((((((((((.((((((...((....))....))))))...(((...((((.(((((....)))))..))))..))).......))))))((((....)))).))))............. ( -48.80) >Blich.0 611619 120 + 4222334/1280-1400 UUCCGCCCUUUAGUGCUGCAGCAAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAU ((((((((...((((((...((....))....))))))((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).))))............. ( -44.20) >Banth.0 245805 120 + 5227293/1280-1400 UUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAU ((((((((...((((((...((....))....))))))((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).))))............. ( -44.00) >Bcere.0 279198 120 + 5224283/1280-1400 UUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAU ((((((((...((((((...((....))....))))))((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).))))............. ( -44.00) >Bhalo.0 558855 120 + 4202352/1280-1400 UCGAUGCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCAGUGGAGCAUGUGGUUUAAU .....(((((((((((..((((....)).))..)))))...(((...((((.(((((....)))))..))))..))).......))))))...((((.((......)).))))....... ( -37.40) >consensus UUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAU ....((((((.((((((...((....))....))))))...(((...((((.(((((....)))))..))))..))).......))))))...((((.((......)).))))....... (-40.90 = -40.46 + -0.44)

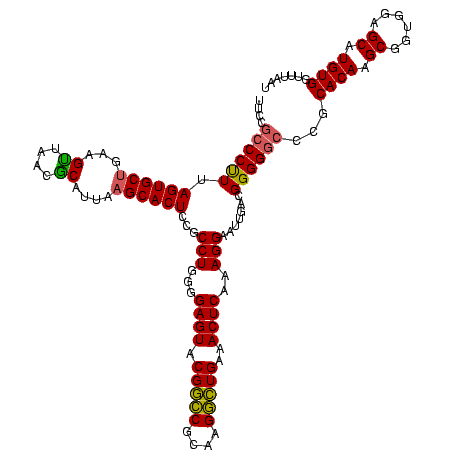
| Location | 634,589 – 634,707 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -45.68 |
| Consensus MFE | -39.52 |
| Energy contribution | -44.40 |
| Covariance contribution | 4.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 118 + 4214630/1440-1560 CUGACAAUCCUAGAGAUAGGACGU-CCCCUUCGGGGG-CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUGA (((....(((((....)))))...-((((....))))-))).....((((.(((((.(((......)))(((((((((....)))((.(((......))))).)))))))).))))))). ( -44.80) >Blich.0 611619 118 + 4222334/1440-1560 CUGACAACCCUAGAGAUAGGGCUU-CCCCUUCGGGGG-CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUGA (((....(((((....)))))...-((((....))))-))).....((((.(((((.(((......)))(((((((((....)))((.(((......))))).)))))))).))))))). ( -46.30) >Banth.0 245805 118 + 5227293/1440-1560 CUGACAACCCUAGAGAUAGGGCUU-CUCCUUCGGGAG-CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUGA (((....(((((....)))))...-((((....))))-))).....((((.(((((.(((......)))(((((((((....)))((.(((......))))).)))))))).))))))). ( -44.20) >Bcere.0 279198 118 + 5224283/1440-1560 CUGACAACCCUAGAGAUAGGGCUU-CUCCUUCGGGAG-CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUGA (((....(((((....)))))...-((((....))))-))).....((((.(((((.(((......)))(((((((((....)))((.(((......))))).)))))))).))))))). ( -44.20) >Bhalo.0 558855 120 + 4202352/1440-1560 UUGACCACCCUAGAGAUAGGGCUUUCCCCUUCGGGGGACAAAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUGA ...(((((((((....))).(((((((((....)))))..))))....)))))).((.((((((.....(((((((((....)))((.(((......))))).))))))))))))..)). ( -48.90) >consensus CUGACAACCCUAGAGAUAGGGCUU_CCCCUUCGGGGG_CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUGA (((....(((((....)))))....((((....)))).))).....((((.(((((.(((......)))(((((((((....)))((.(((......))))).)))))))).))))))). (-39.52 = -44.40 + 4.88)

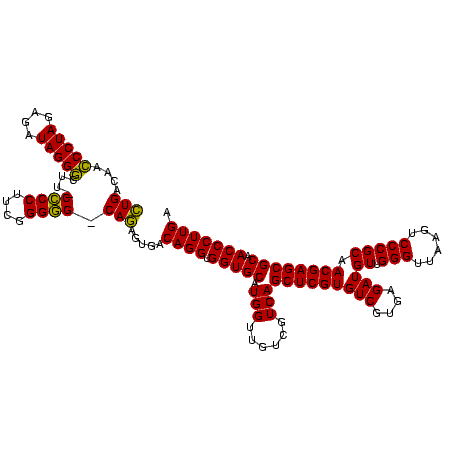
| Location | 634,589 – 634,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.33 |
| Mean single sequence MFE | -41.02 |
| Consensus MFE | -42.92 |
| Energy contribution | -38.76 |
| Covariance contribution | -4.16 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.66 |
| Structure conservation index | 1.05 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 120 + 4214630/1680-1800 UGGACAGAACAAAGGGCAGCGAAACCGCGAGGUUAAGCCAAUCCCACAAAUCUGUUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGCUGGAAUCGCUAGUAAUCGCGGAUC .(((((((.....(((..((..((((....))))..))....))).....)))))))........((((((((((........))))))((((.(((((.....)))))..)))).)))) ( -41.90) >Blich.0 611619 120 + 4222334/1680-1800 UGGGCAGAACAAAGGGCAGCGAAGCCGCGAGGCUAAGCCAAUCCCACAAAUCUGUUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGCUGGAAUCGCUAGUAAUCGCGGAUC .(((((((.....(((..((..((((....))))..))....))).....)))))))........((((((((((........))))))((((.(((((.....)))))..)))).)))) ( -43.80) >Banth.0 245805 120 + 5227293/1680-1800 UGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCGCUAGUAAUCGCGGAUC .(((((((.....(((..((...(((....)))...))....))).....)))))))...(((((((((((((((........)))))).....(((((.....))))))))).))))). ( -38.30) >Bcere.0 279198 120 + 5224283/1680-1800 UGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCGCUAGUAAUCGCGGAUC .(((((((.....(((..((...(((....)))...))....))).....)))))))...(((((((((((((((........)))))).....(((((.....))))))))).))))). ( -38.30) >Bhalo.0 558855 120 + 4202352/1680-1800 UGGAUGGUACAAAGGGUUGCGAAGCCGCGAGGUGAAGCCAAUCCCAGAAAGCCAUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAAGCCGGAAUUGCUAGUAAUCGCGGAUC .(((((((.....(((((((...(((....)))...).))))))......)))))))....(((((..(((((((........))))))).....))))).((((.(....).))))... ( -42.80) >consensus UGGACGGUACAAAGGGCUGCGAAACCGCGAGGUGAAGCCAAUCCCACAAAACCGUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAAGCUGGAAUCGCUAGUAAUCGCGGAUC .(((((((.....(((..((...(((....)))...))....))).....)))))))....(((((..(((((((........))))))).....))))).((((........))))... (-42.92 = -38.76 + -4.16)


| Location | 634,589 – 634,708 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.54 |
| Mean single sequence MFE | -41.34 |
| Consensus MFE | -39.36 |
| Energy contribution | -39.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 119 + 4214630/1880-2000 UGAGGUAACCUUUUAGGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUA-C ...((((.((((..(((((((..(((((...))))).....((((((.......)))))).....))((((((((((....))))))))))......)))))....)))).))))...-. ( -41.60) >Blich.0 611619 118 + 4222334/1880-2000 UGAGGUAACCUUUU-GGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUA-C ...((((.((((((-(...((.(((......))).)).)))).((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))).))))...-. ( -41.60) >Banth.0 245805 119 + 5227293/1880-2000 UGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA-U ...(((..((.....)).)))..(((((...)))))..((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..)).........-. ( -41.50) >Bcere.0 279198 118 + 5224283/1880-2000 UGGGGUAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA-U ...(((..((....-)).)))..(((((...)))))..((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..)).........-. ( -41.50) >Bhalo.0 558855 119 + 4202352/1880-2000 UGGGGUAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUACGGAGUUUUUAAC .(((((.(((((((-(...((.(((......))).)).)))((((((.......))))))...)))))(((((((((....))))))))).....)))))..(((....)))........ ( -40.50) >consensus UGGGGUAACCUUUU_GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAGAUUUUA_C ...(((..((.....)).)))..(((((...))))).......((..(((((((.((....))....((((((((((....))))))))))..))))).))..))............... (-39.36 = -39.36 + -0.00)

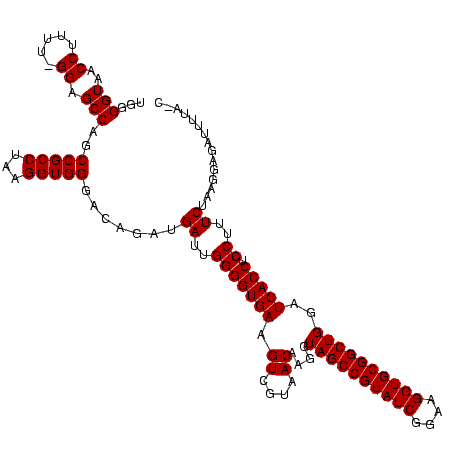
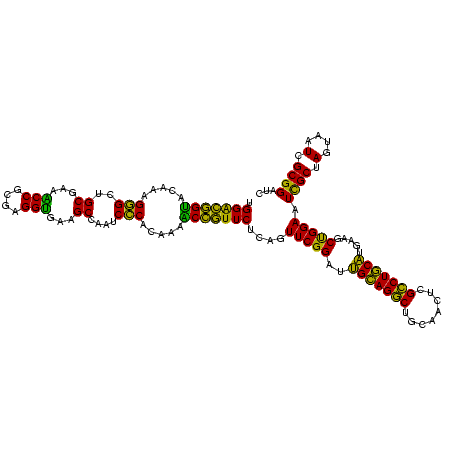
| Location | 634,589 – 634,707 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.60 |
| Mean single sequence MFE | -44.90 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.81 |
| Structure conservation index | 0.66 |
| SVM decision value | 4.19 |
| SVM RNA-class probability | 0.999829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 118 + 4214630/1920-2040 GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUA-CGGAAUAUAAGACCUUG-GGUCUUAUAAACAGAACGUUCCC .......((..((...(((((....((((((((((((....))))))))).....)))((((.....)))).....))-)))..(((((((((...-))))))))).......))..)). ( -40.80) >Blich.0 611619 119 + 4222334/1920-2040 GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUA-CGGAAUAUAAGACCUUGCGGUCUUAUAGACAGGUGCGUUUG ...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))...........((-(.(..(((((((((....)))))))))..)..)))...... ( -41.90) >Banth.0 245805 113 + 5227293/1920-2040 GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA-UG------AACGCUGUUCAUCAAUAUAAGUUUCCGUGUUUC ...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))(((((((((((((-((------(((...)))))))))).....)))))))).... ( -45.80) >Bcere.0 279198 113 + 5224283/1920-2040 GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA-UG------AACGCUGUUCAUCAAUAUAAGUUUCCGUGUUUC ...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))(((((((((((((-((------(((...)))))))))).....)))))))).... ( -45.80) >Bhalo.0 558855 120 + 4202352/1920-2040 GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUACGGAGUUUUUAACUCUAGUCGACCAUUCGCAGAUUCAUCUGCGGAAGGACGCU ..((((((((((((..(((((..((((.(((((((((....)))))))))((....)).))))..)))))..))....)))))))))).((.((((((((....)))))))).))..... ( -50.20) >consensus GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAGAUUUUA_CG__A___AACACUUUGCAGCAUUAUAAGCAGAAGGGUUUC ...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))....................................................... (-29.70 = -29.70 + -0.00)

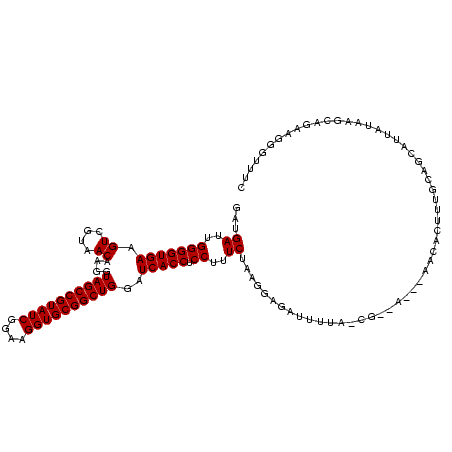
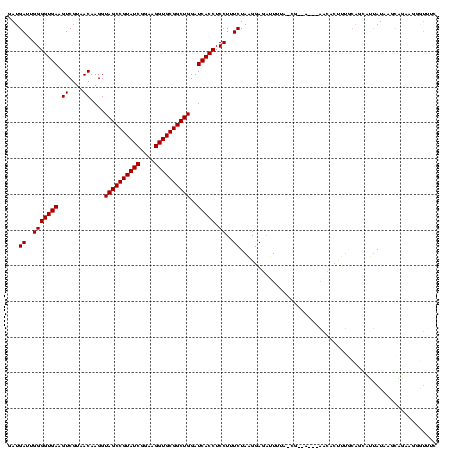
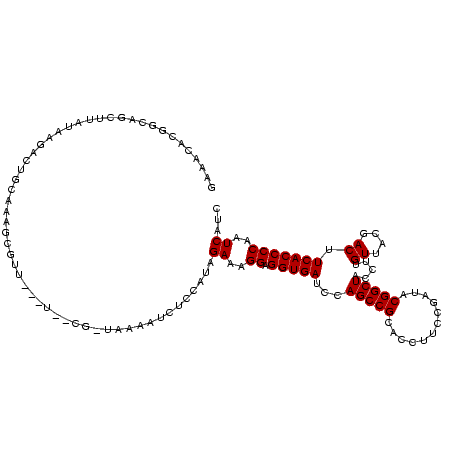
| Location | 634,589 – 634,707 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.60 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -15.20 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 118 + 4214630/1920-2040 GGGAACGUUCUGUUUAUAAGACC-CAAGGUCUUAUAUUCCG-UAUAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC (((........(..(((((((((-...)))))))))..)((-((.....((((.....))))(((.....(((((............))))))))....)))).......)))....... ( -28.40) >Blich.0 611619 119 + 4222334/1920-2040 CAAACGCACCUGUCUAUAAGACCGCAAGGUCUUAUAUUCCG-UAUAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC ...........((((((((((((....)))))))))..(((-(((....((((.....))))((((.........)))).....))))))............)))............... ( -30.90) >Banth.0 245805 113 + 5227293/1920-2040 GAAACACGGAAACUUAUAUUGAUGAACAGCGUU------CA-UCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC .......(((.......((((((((((...)))------))-)))))..)))...((..((.(((((...(((((............))))).....((....)).)))))))..))... ( -26.80) >Bcere.0 279198 113 + 5224283/1920-2040 GAAACACGGAAACUUAUAUUGAUGAACAGCGUU------CA-UCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC .......(((.......((((((((((...)))------))-)))))..)))...((..((.(((((...(((((............))))).....((....)).)))))))..))... ( -26.80) >Bhalo.0 558855 120 + 4202352/1920-2040 AGCGUCCUUCCGCAGAUGAAUCUGCGAAUGGUCGACUAGAGUUAAAAACUCCGUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC ((((.((...((((((....))))))...)).)).)).((((.....))))(((((.((((.((....))(((((............))))).)))).)))))................. ( -33.20) >consensus GAAACACGGCAGCUUAUAAGACUGCAAAGCGUU___U__CG_UAAAAUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC .......................................................((..((.(((((...(((((............))))).....((....)).)))))))..))... (-15.20 = -15.20 + 0.00)

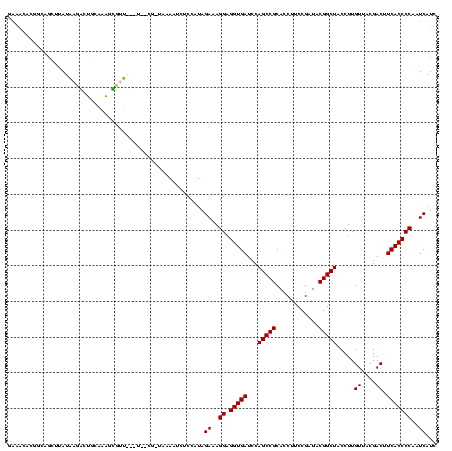
| Location | 634,589 – 634,702 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.01 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -13.98 |
| Energy contribution | -12.58 |
| Covariance contribution | -1.40 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.42 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.987027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
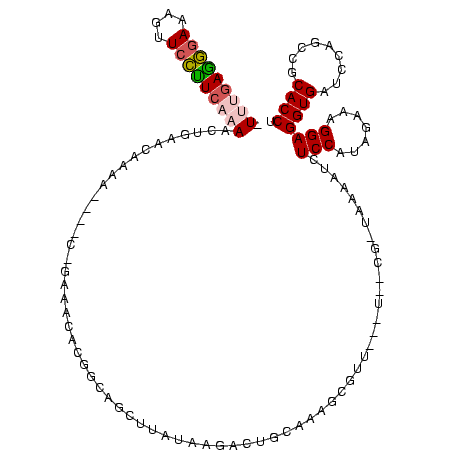
>Bsubt.0 634589 113 + 4214630/1960-2080 AGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUA-CGGAAUAUAAGACCUUG-GGUCUUAUAAACAGAACGUUCCCUG----UCUUGUUUAGUUUUGAAGGAUCAUUCCUUCGAA- .((.(((.(((.......((((.....)))).......-.....(((((((((...-)))))))))..)))..))).))...----...........(((((((((....)))))))))- ( -29.90) >Blich.0 611619 114 + 4222334/1960-2080 AGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUA-CGGAAUAUAAGACCUUGCGGUCUUAUAGACAGGUGCGUUUGGA----UCUUGUUUAGUUUUGAAGGAUCAUUCCUUCAAA- ......(((..(((((((((((.....)))).......-.(...(((((((((....)))))))))..).)))).)))..).----)).........(((((((((....)))))))))- ( -34.50) >Banth.0 245805 108 + 5227293/1960-2080 AGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA-UG------AACGCUGUUCAUCAAUAUAAGUUUCCGUGUUUC-G----UUUUGUUCAGUUUUGAGAGAACUAUCUCUCAUAU ......(((((((((((((((.......)))).(((((-((------(((...))))))))))..........)))....-.----....)))))))).(((((((....)))))))... ( -30.60) >Bcere.0 279198 108 + 5224283/1960-2080 AGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA-UG------AACGCUGUUCAUCAAUAUAAGUUUCCGUGUUUC-G----UUUUGUUCAGUUUUGAGAGAACUAUCUCUCAUAU ......(((((((((((((((.......)))).(((((-((------(((...))))))))))..........)))....-.----....)))))))).(((((((....)))))))... ( -30.60) >Bhalo.0 558855 120 + 4202352/1960-2080 AGGUGCGGCUGGAUCACCUCCUUUCUACGGAGUUUUUAACUCUAGUCGACCAUUCGCAGAUUCAUCUGCGGAAGGACGCUUGAAGCUUUUGUUCAGUUUUGAAGGAAUUGUUCCUUCAAA .(((.((((((((....((((.......))))........))))))))))).((((((((....)))))))).((((((.....))....))))...(((((((((.....))))))))) ( -40.60) >consensus AGGUGCGGCUGGAUCACCUCCUUUCUAAGGAGAUUUUA_CG__A___AACACUUUGCAGCAUUAUAAGCAGAAGGGUUUC_G____UUUUGUUCAGUUUUGAAGGAACUAUCCCUCAAA_ .......(((((((...((((.......))))............................((.......))...................)))))))(((((((((....))))))))). (-13.98 = -12.58 + -1.40)

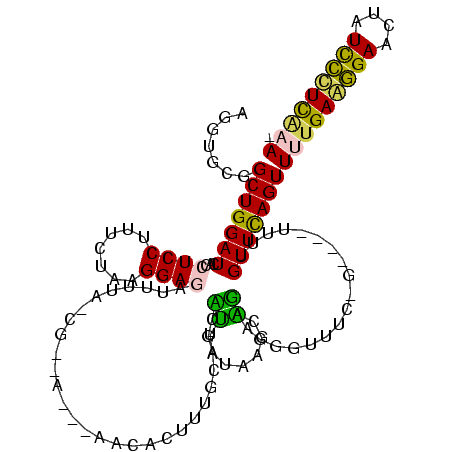
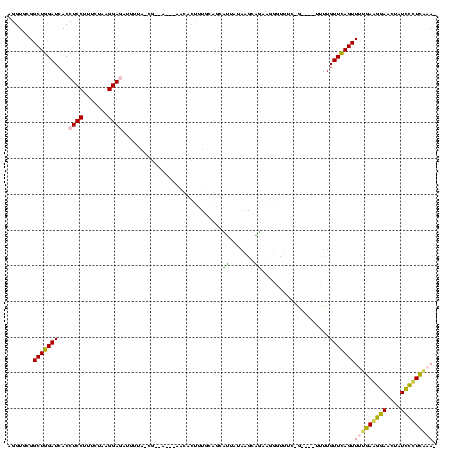
| Location | 634,589 – 634,702 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.01 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -12.64 |
| Energy contribution | -12.88 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.41 |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.998671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 113 + 4214630/1960-2080 -UUCGAAGGAAUGAUCCUUCAAAACUAAACAAGA----CAGGGAACGUUCUGUUUAUAAGACC-CAAGGUCUUAUAUUCCG-UAUAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCU -...((((((....)))))).....((((((..(----(.(....)))..))))))(((((((-...))))))).......-.......((((.....))))((((.........)))). ( -28.60) >Blich.0 611619 114 + 4222334/1960-2080 -UUUGAAGGAAUGAUCCUUCAAAACUAAACAAGA----UCCAAACGCACCUGUCUAUAAGACCGCAAGGUCUUAUAUUCCG-UAUAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCU -(((((((((....)))))))))...........----........((((....(((((((((....))))))))).....-.......((((.....)))))))).............. ( -33.20) >Banth.0 245805 108 + 5227293/1960-2080 AUAUGAGAGAUAGUUCUCUCAAAACUGAACAAAA----C-GAAACACGGAAACUUAUAUUGAUGAACAGCGUU------CA-UCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCU ...(((((((....))))))).............----.-.......(((.......((((((((((...)))------))-)))))(((((.......)))))...))).......... ( -27.80) >Bcere.0 279198 108 + 5224283/1960-2080 AUAUGAGAGAUAGUUCUCUCAAAACUGAACAAAA----C-GAAACACGGAAACUUAUAUUGAUGAACAGCGUU------CA-UCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCU ...(((((((....))))))).............----.-.......(((.......((((((((((...)))------))-)))))(((((.......)))))...))).......... ( -27.80) >Bhalo.0 558855 120 + 4202352/1960-2080 UUUGAAGGAACAAUUCCUUCAAAACUGAACAAAAGCUUCAAGCGUCCUUCCGCAGAUGAAUCUGCGAAUGGUCGACUAGAGUUAAAAACUCCGUAGAAAGGAGGUGAUCCAGCCGCACCU (((((((((.....))))))))).(((......(((((..((((.((...((((((....))))))...)).)).)).))))).....((((.......))))......)))........ ( -35.10) >consensus _UUUGAGGGAAAGUUCCUUCAAAACUGAACAAAA____C_GAAACACGGCAGCUUAUAAGACUGCAAAGCGUU___U__CG_UAAAAUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCU .(((((((((....)))))))))..................................................................(((.......)))((((.........)))). (-12.64 = -12.88 + 0.24)

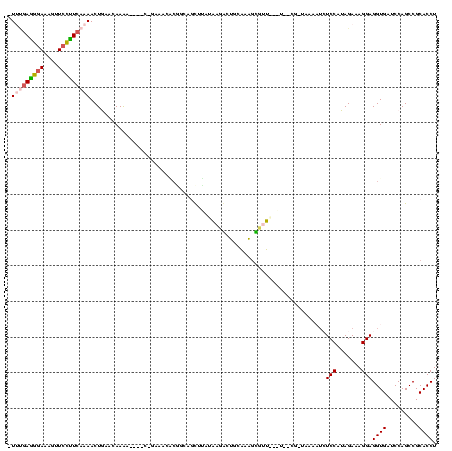
| Location | 634,589 – 634,696 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 59.95 |
| Mean single sequence MFE | -31.46 |
| Consensus MFE | -10.68 |
| Energy contribution | -10.64 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.38 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.34 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 107 + 4214630/1977-2097 CUCCUUUCUAAGGAUAUUAUA-CGGAAUAUAAGACCUUG-GGUCUUAUAAACAGAACGUUCCCUG----UCUUGUUUAGUUUUGAAGGAUCAUUCCUUCGAA-------ACGUGUUCUUU .((((.....)))).......-.(((((((((((((...-)))))).((((((..(((.....))----)..))))))(((((((((((....)))))))))-------))))))))).. ( -33.50) >Blich.0 611619 108 + 4222334/1977-2097 CUCCUUUCUAAGGAUAUUAUA-CGGAAUAUAAGACCUUGCGGUCUUAUAGACAGGUGCGUUUGGA----UCUUGUUUAGUUUUGAAGGAUCAUUCCUUCAAA-------GCGUGUUCUUU .((((.....)))).......-.(((((((((((((....)))))).((((((((..(.....).----.))))))))(((((((((((....)))))))))-------))))))))).. ( -35.80) >Banth.0 245805 108 + 5227293/1977-2097 CUCCUUUCUAUGGAGAAUUGA-UG------AACGCUGUUCAUCAAUAUAAGUUUCCGUGUUUC-G----UUUUGUUCAGUUUUGAGAGAACUAUCUCUCAUAUAUAAAUGUAUGUUCUUU ........(((((((((((((-((------(((...)))))))))).....))))))))....-.----..............((((((....))))))((((((....))))))..... ( -24.90) >Bcere.0 279198 108 + 5224283/1977-2097 CUCCUUUCUAUGGAGAAUUGA-UG------AACGCUGUUCAUCAAUAUAAGUUUCCGUGUUUC-G----UUUUGUUCAGUUUUGAGAGAACUAUCUCUCAUAUAUAAAUGUAUGUUCUUU ........(((((((((((((-((------(((...)))))))))).....))))))))....-.----..............((((((....))))))((((((....))))))..... ( -24.90) >Bhalo.0 558855 120 + 4202352/1977-2097 CUCCUUUCUACGGAGUUUUUAACUCUAGUCGACCAUUCGCAGAUUCAUCUGCGGAAGGACGCUUGAAGCUUUUGUUCAGUUUUGAAGGAAUUGUUCCUUCAAAACAACUGAUUGUUUCUU .(((((((...(((((.....)))))............(((((....)))))))))))).....(((((..(((((..(((((((((((.....)))))))))))))).))..))))).. ( -38.20) >consensus CUCCUUUCUAAGGAGAUUUUA_CG__A___AACACUUUGCAGCAUUAUAAGCAGAAGGGUUUC_G____UUUUGUUCAGUUUUGAAGGAACUAUCCCUCAAA_A_AA_UGCAUGUUCUUU ((((.......)))).................................................................(((((((((....))))))))).................. (-10.68 = -10.64 + -0.04)

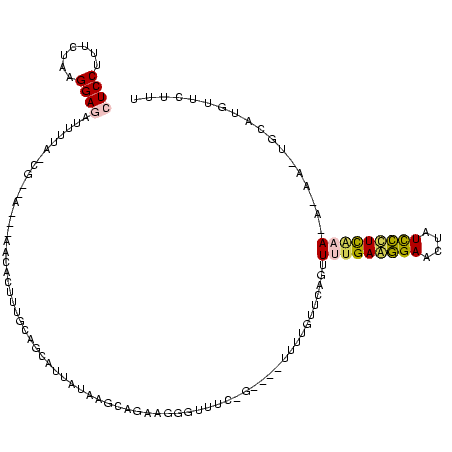
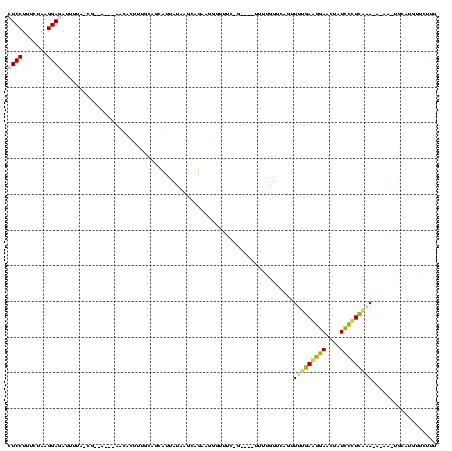
| Location | 634,589 – 634,696 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 59.95 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -9.06 |
| Energy contribution | -9.70 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.77 |
| Structure conservation index | 0.32 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 634589 107 + 4214630/1977-2097 AAAGAACACGU-------UUCGAAGGAAUGAUCCUUCAAAACUAAACAAGA----CAGGGAACGUUCUGUUUAUAAGACC-CAAGGUCUUAUAUUCCG-UAUAAUAUCCUUAGAAAGGAG ..(((((..((-------((.((((((....)))))).)))).........----..(....))))))(..(((((((((-...)))))))))..)..-.......((((.....)))). ( -28.70) >Blich.0 611619 108 + 4222334/1977-2097 AAAGAACACGC-------UUUGAAGGAAUGAUCCUUCAAAACUAAACAAGA----UCCAAACGCACCUGUCUAUAAGACCGCAAGGUCUUAUAUUCCG-UAUAAUAUCCUUAGAAAGGAG .......(((.-------(((((((((....)))))))))...........----................(((((((((....)))))))))...))-)......((((.....)))). ( -28.80) >Banth.0 245805 108 + 5227293/1977-2097 AAAGAACAUACAUUUAUAUAUGAGAGAUAGUUCUCUCAAAACUGAACAAAA----C-GAAACACGGAAACUUAUAUUGAUGAACAGCGUU------CA-UCAAUUCUCCAUAGAAAGGAG ....................(((((((....))))))).............----.-.......(....)....((((((((((...)))------))-))))).((((.......)))) ( -23.70) >Bcere.0 279198 108 + 5224283/1977-2097 AAAGAACAUACAUUUAUAUAUGAGAGAUAGUUCUCUCAAAACUGAACAAAA----C-GAAACACGGAAACUUAUAUUGAUGAACAGCGUU------CA-UCAAUUCUCCAUAGAAAGGAG ....................(((((((....))))))).............----.-.......(....)....((((((((((...)))------))-))))).((((.......)))) ( -23.70) >Bhalo.0 558855 120 + 4202352/1977-2097 AAGAAACAAUCAGUUGUUUUGAAGGAACAAUUCCUUCAAAACUGAACAAAAGCUUCAAGCGUCCUUCCGCAGAUGAAUCUGCGAAUGGUCGACUAGAGUUAAAAACUCCGUAGAAAGGAG .........((((((...((((((((.....)))))))))))))).....(((((..((((.((...((((((....))))))...)).)).)).))))).....((((.......)))) ( -36.80) >consensus AAAGAACACACA_UU_U_UUUGAGGGAAAGUUCCUUCAAAACUGAACAAAA____C_GAAACACGGCAGCUUAUAAGACUGCAAAGCGUU___U__CG_UAAAAUCUCCAUAGAAAGGAG ..................(((((((((....))))))))).................................................................((((.......)))) ( -9.06 = -9.70 + 0.64)

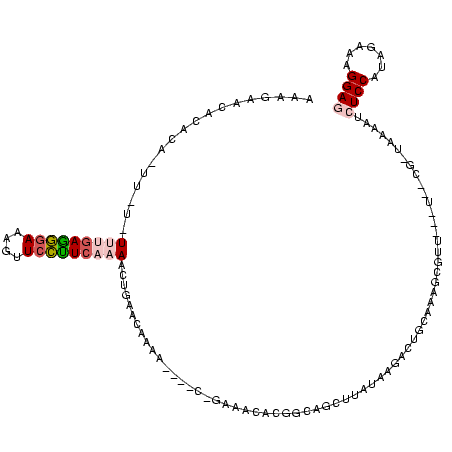
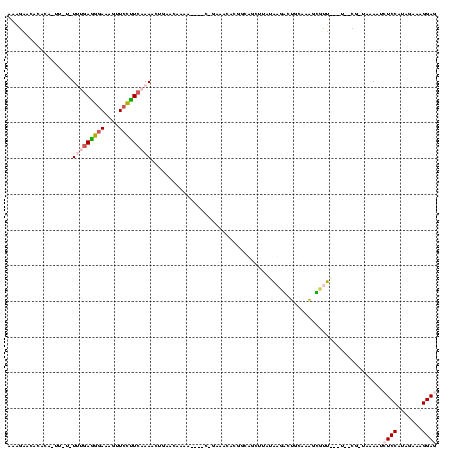
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:22:00 2006