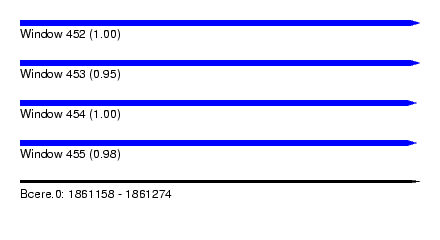
| Sequence ID | Bcere.0 |
|---|---|
| Location | 1,861,158 – 1,861,274 |
| Length | 116 |
| Max. P | 0.999618 |

| Location | 1,861,158 – 1,861,274 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -31.02 |
| Energy contribution | -29.70 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 1861158 116 + 5224283/40-160 GGUGUUUACAUGGCGAUUACAAUUCAAUUGCCGGACACUGCAGCGUCUCAUGCAAAGCCAAGUAUGGAAAUGGCGAUACUUUGCGUGAGGUAGACAAGUAAUAUACAAU----CGCCUAA (((((((((.((((((((.......)))))))).............(((((((((((....((((....)).))....)))))))))))))))))..(((...)))...----.)))... ( -33.40) >Banth.0 1723710 120 + 5227293/40-160 GGUGUUUAAAUGGCGAUUACAAUUCAAUUGCCGGACACUGCAGCGUCUCAUGCAAAGCCAAGUAUGGAAAUGGCGAUACUUUGCGUGAGGUAGACAAGUAAUAUACAAUUCAUCGCCAAA ((((((....((((((((.......))))))))))))))...(((.(((((((((((....((((....)).))....)))))))))))(((...........))).......))).... ( -32.40) >Bsubt.0 2748265 114 + 4214630/40-160 GGUGAUGUAAUGGCAAGCAAGAUGAGAAUGCCAGAUUCCGCAACGUCCCAUGCAAAGCCUACUAUUGGGAAGGCGAUACUUUGCAUGGGGUAGACU--UUCUAAAAGAU----CGCCCAA ((((((.(..(((((..(.......)..)))))............(((((((((((((((.((....)).)))).....)))))))))))......--.......).))----))))... ( -35.50) >consensus GGUGUUUAAAUGGCGAUUACAAUUCAAUUGCCGGACACUGCAGCGUCUCAUGCAAAGCCAAGUAUGGAAAUGGCGAUACUUUGCGUGAGGUAGACAAGUAAUAUACAAU____CGCCAAA ((((......((((((((.......))))))))............(((((((((((((((..........)))).....))))))))))).......................))))... (-31.02 = -29.70 + -1.32)

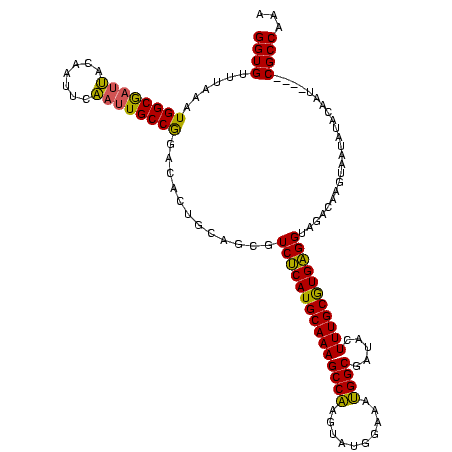
| Location | 1,861,158 – 1,861,274 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -23.65 |
| Energy contribution | -22.10 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 1861158 116 + 5224283/40-160 UUAGGCG----AUUGUAUAUUACUUGUCUACCUCACGCAAAGUAUCGCCAUUUCCAUACUUGGCUUUGCAUGAGACGCUGCAGUGUCCGGCAAUUGAAUUGUAAUCGCCAUGUAAACACC ...((((----((((((..(((.(((((....(((.(((((.....((((..........))))))))).)))(((((....))))).))))).)))..))))))))))........... ( -33.50) >Banth.0 1723710 120 + 5227293/40-160 UUUGGCGAUGAAUUGUAUAUUACUUGUCUACCUCACGCAAAGUAUCGCCAUUUCCAUACUUGGCUUUGCAUGAGACGCUGCAGUGUCCGGCAAUUGAAUUGUAAUCGCCAUUUAAACACC ..(((((((.....((...(((.(((((....(((.(((((.....((((..........))))))))).)))(((((....))))).))))).)))))....))))))).......... ( -29.80) >Bsubt.0 2748265 114 + 4214630/40-160 UUGGGCG----AUCUUUUAGAA--AGUCUACCCCAUGCAAAGUAUCGCCUUCCCAAUAGUAGGCUUUGCAUGGGACGUUGCGGAAUCUGGCAUUCUCAUCUUGCUUGCCAUUACAUCACC ..(((((----((....(((..--...))).((((((((((((...((..........))..))))))))))))..)))))......(((((..(.......)..)))))........)) ( -31.10) >consensus UUAGGCG____AUUGUAUAUUACUUGUCUACCUCACGCAAAGUAUCGCCAUUUCCAUACUUGGCUUUGCAUGAGACGCUGCAGUGUCCGGCAAUUGAAUUGUAAUCGCCAUUUAAACACC ...((((........................((((.(((((.....((((..........))))))))).))))..((((.......))))..............))))........... (-23.65 = -22.10 + -1.55)

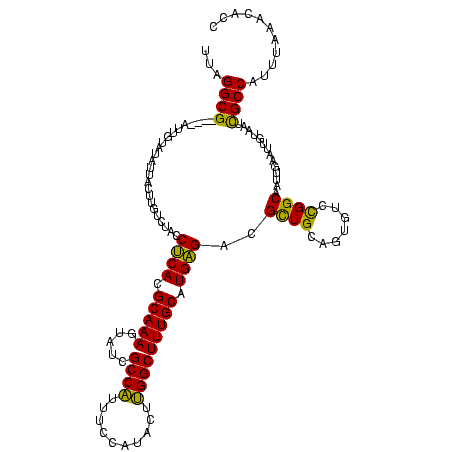
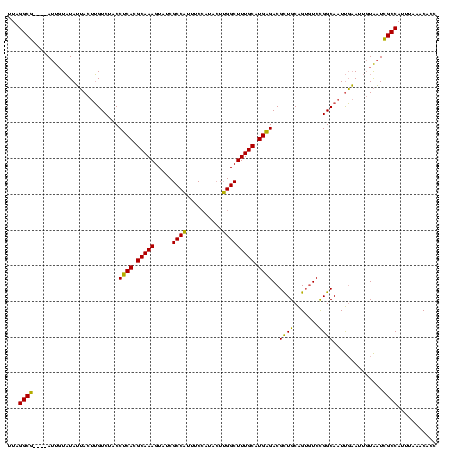
| Location | 1,861,158 – 1,861,273 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -38.13 |
| Consensus MFE | -26.61 |
| Energy contribution | -26.73 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.70 |
| SVM decision value | 3.79 |
| SVM RNA-class probability | 0.999618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 1861158 115 + 5224283/80-200 CAGCGUCUCAUGCAAAGCCAAGUAUGGAAAUGGCGAUACUUUGCGUGAGGUAGACAAGUAAUAUACAAU----CGCCUAAAUGAGCUCGUAUCAUUUUUUAUAUU-UGGCUUUGCACCUU ..........(((((((((((((((((((..(((((((((((....)))))).....(((...)))..)----))))..(((((.......))))))))))))))-)))))))))).... ( -38.00) >Banth.0 1723710 119 + 5227293/80-200 CAGCGUCUCAUGCAAAGCCAAGUAUGGAAAUGGCGAUACUUUGCGUGAGGUAGACAAGUAAUAUACAAUUCAUCGCCAAAAUGAGCUCGUAUCAUUUUUUGUAUU-UGGCUUUGCACCUU ..........((((((((((((((..(((.((((((((((((....)))))).....(((...)))......)))))).(((((.......))))))))..))))-)))))))))).... ( -39.40) >Bsubt.0 2748265 107 + 4214630/80-200 CAACGUCCCAUGCAAAGCCUACUAUUGGGAAGGCGAUACUUUGCAUGGGGUAGACU--UUCUAAAAGAU----CGCCCAAGUAAGCU-------UUUCUGAUAUAGUGGCUUUGCACCUU ..........(((((((((.(((((.((((((((..(((((......((((.((((--(.....))).)----)))))))))).)))-------)))))...)))))))))))))).... ( -37.00) >consensus CAGCGUCUCAUGCAAAGCCAAGUAUGGAAAUGGCGAUACUUUGCGUGAGGUAGACAAGUAAUAUACAAU____CGCCAAAAUGAGCUCGUAUCAUUUUUUAUAUU_UGGCUUUGCACCUU ..........((((((((((((((((((((.((((.((((((....)))))).....(((...))).......))))...((((.......)))))))))))))).)))))))))).... (-26.61 = -26.73 + 0.12)

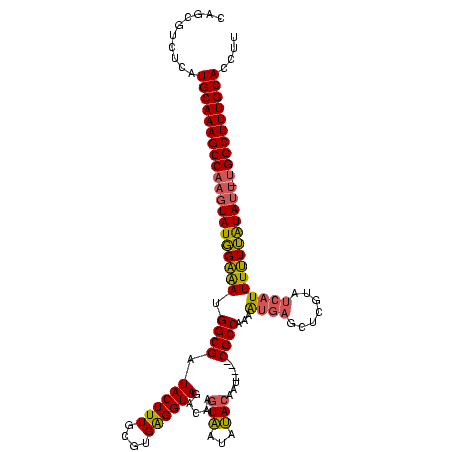
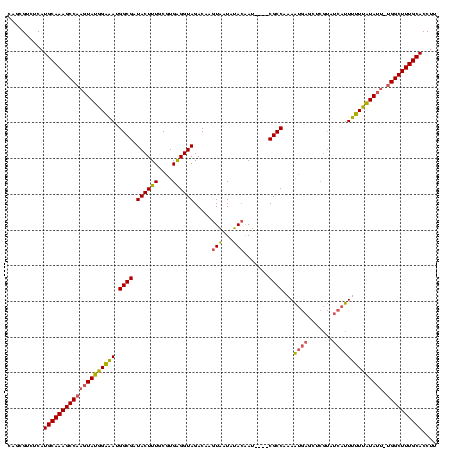
| Location | 1,861,158 – 1,861,273 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -20.19 |
| Energy contribution | -19.31 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 1861158 115 + 5224283/80-200 AAGGUGCAAAGCCA-AAUAUAAAAAAUGAUACGAGCUCAUUUAGGCG----AUUGUAUAUUACUUGUCUACCUCACGCAAAGUAUCGCCAUUUCCAUACUUGGCUUUGCAUGAGACGCUG ...(((((((((((-(.(((..(((.((((((..((.....((((((----(.((.....)).)))))))......))...))))))...)))..))).))))))))))))......... ( -31.00) >Banth.0 1723710 119 + 5227293/80-200 AAGGUGCAAAGCCA-AAUACAAAAAAUGAUACGAGCUCAUUUUGGCGAUGAAUUGUAUAUUACUUGUCUACCUCACGCAAAGUAUCGCCAUUUCCAUACUUGGCUUUGCAUGAGACGCUG ...(((((((((((-(......(((((((.......)))))))(((((((..((((....................))))..)))))))..........))))))))))))......... ( -34.55) >Bsubt.0 2748265 107 + 4214630/80-200 AAGGUGCAAAGCCACUAUAUCAGAAA-------AGCUUACUUGGGCG----AUCUUUUAGAA--AGUCUACCCCAUGCAAAGUAUCGCCUUCCCAAUAGUAGGCUUUGCAUGGGACGUUG ...(((((((((((((((.......(-------((....)))(((((----(((((((((..--...)))........)))).))))))).....))))).))))))))))......... ( -31.30) >consensus AAGGUGCAAAGCCA_AAUAUAAAAAAUGAUACGAGCUCAUUUAGGCG____AUUGUAUAUUACUUGUCUACCUCACGCAAAGUAUCGCCAUUUCCAUACUUGGCUUUGCAUGAGACGCUG ..(((.....)))....................(((.....(((((...................))))).((((.(((((.....((((..........))))))))).))))..))). (-20.19 = -19.31 + -0.88)

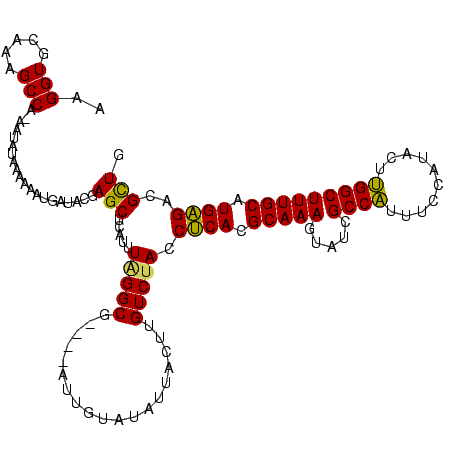
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:21:31 2006