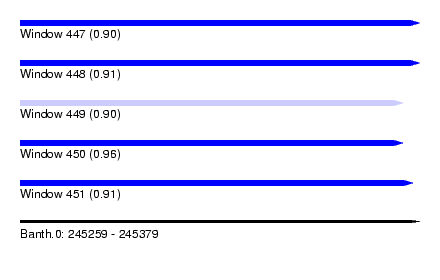
| Sequence ID | Banth.0 |
|---|---|
| Location | 245,259 – 245,379 |
| Length | 120 |
| Max. P | 0.963257 |

| Location | 245,259 – 245,379 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -37.41 |
| Consensus MFE | -35.59 |
| Energy contribution | -34.49 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.900993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 245259 120 + 5227293/0-120 ACUUCGGAGGAUUAGCUCAGCUGGGAGAGCACCUGCCUUACAAGCAGGGGGUCGGCGGUUCGAUCCCGUCAUCCUCCACCAUAAUAUUAUAAUGAAAAGUAAAAUGAUAUUAAUGUAAAG .....(((((((..((((........))))..((((.......))))((((((((....))))))))...)))))))....(((((((((.............)))))))))........ ( -37.02) >Bcere.0 278654 120 + 5224283/0-120 ACUUCGGAGGAUUAGCUCAGCUGGGAGAGCACCUGCCUUACAAGCAGGGGGUCGGCGGUUCGAUCCCGUCAUCCUCCACCAUAAUAUUAUAAUGAAAAGUAAAAUGACAUUAAUGUAAAG .....(((((((..((((........))))..((((.......))))((((((((....))))))))...)))))))............(((((.............)))))........ ( -33.92) >Bhalo.0 558270 120 + 4202352/0-120 CAUUCGGAGGAUUAGCUCAGCUGGGAGAGCACCUGCCUUACAAGCAGGGGGUCGGCGGUUCGAUCCCGUCAUCCUCCACCAUUUUAUUGGUCCGGUAGUUCAGUUGGUUAGAAUGCCUGC ((((((((((((..((((........))))..((((.......))))((((((((....))))))))...)))))))((((......))))((((........))))...)))))..... ( -41.30) >consensus ACUUCGGAGGAUUAGCUCAGCUGGGAGAGCACCUGCCUUACAAGCAGGGGGUCGGCGGUUCGAUCCCGUCAUCCUCCACCAUAAUAUUAUAAUGAAAAGUAAAAUGAUAUUAAUGUAAAG .....(((((((..((((........))))..((((.......))))((((((((....))))))))...)))))))....(((((((((.............)))))))))........ (-35.59 = -34.49 + -1.10)

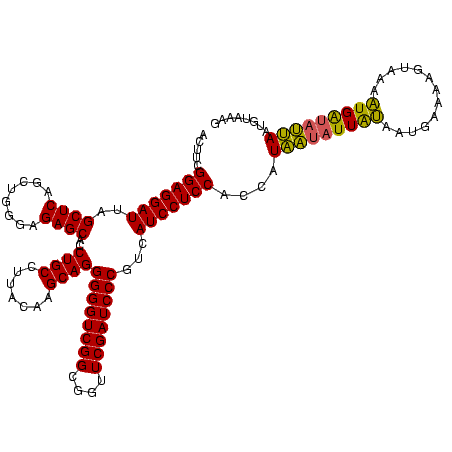
| Location | 245,259 – 245,379 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -33.40 |
| Energy contribution | -33.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 245259 120 + 5227293/0-120 CUUUACAUUAAUAUCAUUUUACUUUUCAUUAUAAUAUUAUGGUGGAGGAUGACGGGAUCGAACCGCCGACCCCCUGCUUGUAAGGCAGGUGCUCUCCCAGCUGAGCUAAUCCUCCGAAGU .......................((((((((((....))))))(((((((...(((.(((......))))))((((((.....)))))).((((........))))..))))))))))). ( -35.30) >Bcere.0 278654 120 + 5224283/0-120 CUUUACAUUAAUGUCAUUUUACUUUUCAUUAUAAUAUUAUGGUGGAGGAUGACGGGAUCGAACCGCCGACCCCCUGCUUGUAAGGCAGGUGCUCUCCCAGCUGAGCUAAUCCUCCGAAGU .......................((((((((((....))))))(((((((...(((.(((......))))))((((((.....)))))).((((........))))..))))))))))). ( -35.30) >Bhalo.0 558270 120 + 4202352/0-120 GCAGGCAUUCUAACCAACUGAACUACCGGACCAAUAAAAUGGUGGAGGAUGACGGGAUCGAACCGCCGACCCCCUGCUUGUAAGGCAGGUGCUCUCCCAGCUGAGCUAAUCCUCCGAAUG .....(((((.......(((......)))((((......))))(((((((...(((.(((......))))))((((((.....)))))).((((........))))..)))))))))))) ( -40.70) >consensus CUUUACAUUAAUAUCAUUUUACUUUUCAUUAUAAUAUUAUGGUGGAGGAUGACGGGAUCGAACCGCCGACCCCCUGCUUGUAAGGCAGGUGCUCUCCCAGCUGAGCUAAUCCUCCGAAGU ..........................................((((((((...(((.(((......))))))((((((.....)))))).((((........))))..)))))))).... (-33.40 = -33.40 + -0.00)

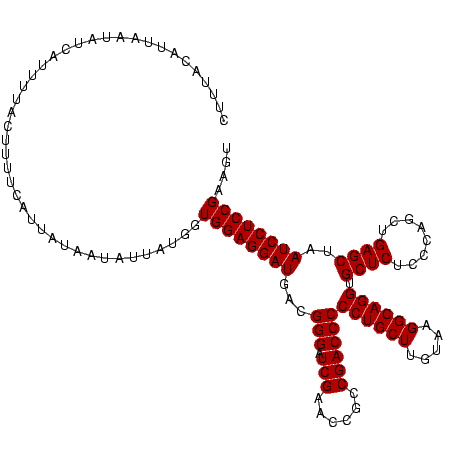
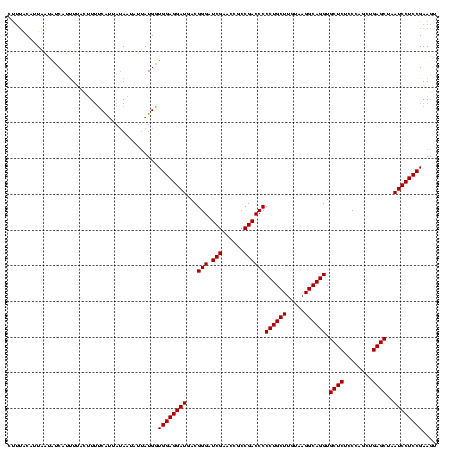
| Location | 245,259 – 245,374 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.58 |
| Mean single sequence MFE | -40.27 |
| Consensus MFE | -24.82 |
| Energy contribution | -22.83 |
| Covariance contribution | -1.99 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 245259 115 + 5227293/320-440 CGGUAAGGCAACGGACUUU---GACUCCGUCAUGCGCUGGUUCGAAUCCAGCUAGCCCAGCCAUUUACGA-GCCAUUAGCUCAGUUGG-UAGAGCAUCUGACUUUUAAUCAGAGGGUCGA .(((..(((.(((((....---...))))).....(((((.......)))))..)))..))).....(((-.((....((((......-..)))).(((((.......))))))).))). ( -39.90) >Bcere.0 278654 115 + 5224283/320-440 CGGUAAGGCAACGGACUUU---GACUCCGUCAUGCGCUGGUUCGAAUCCAGCUAGCCCAGCCAUUUACGA-GCCAUUAGCUCAGUUGG-UAGAGCAUCUGACUUUUAAUCAGAGGGUCGA .(((..(((.(((((....---...))))).....(((((.......)))))..)))..))).....(((-.((....((((......-..)))).(((((.......))))))).))). ( -39.90) >Bhalo.0 558270 120 + 4202352/320-440 CGCUAGGUUGAGGGCCUAGUGGGGGUUGACCCCGUGGAGGUUCGAGUCCUCUCAACCGCAUCGUAUAAAGCGCCCGUAGCUCAAUUGGAUAGAGCGUUUGACUACGGAUCAAAAGGUUAG ((((((((.....))))))))((((....))))..(((((.......))))).((((((..........))..((((((.((((.((.......)).)))))))))).......)))).. ( -41.00) >consensus CGGUAAGGCAACGGACUUU___GACUCCGUCAUGCGCUGGUUCGAAUCCAGCUAGCCCAGCCAUUUACGA_GCCAUUAGCUCAGUUGG_UAGAGCAUCUGACUUUUAAUCAGAGGGUCGA .(((..(((.............((.....))....(((((.......)))))..)))..))).........(((....((((.........)))).(((((.......))))).)))... (-24.82 = -22.83 + -1.99)

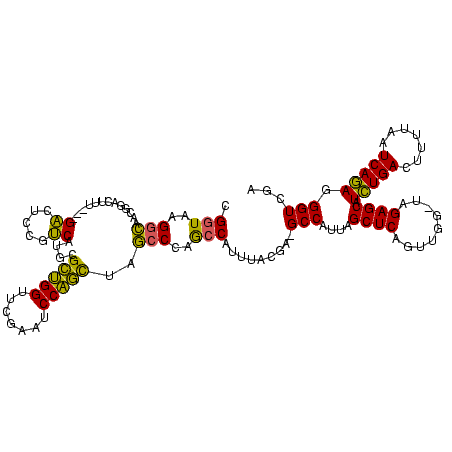
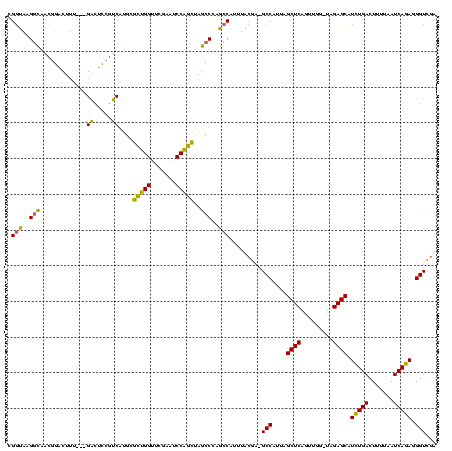
| Location | 245,259 – 245,374 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.58 |
| Mean single sequence MFE | -37.61 |
| Consensus MFE | -23.20 |
| Energy contribution | -23.77 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 245259 115 + 5227293/320-440 UCGACCCUCUGAUUAAAAGUCAGAUGCUCUA-CCAACUGAGCUAAUGGC-UCGUAAAUGGCUGGGCUAGCUGGAUUCGAACCAGCGCAUGACGGAGUC---AAAGUCCGUUGCCUUACCG .(((.((((((((.....)))))).((((..-......))))....)).-))).....((..((((..(((((.......)))))....((((((...---....))))))))))..)). ( -39.10) >Bcere.0 278654 115 + 5224283/320-440 UCGACCCUCUGAUUAAAAGUCAGAUGCUCUA-CCAACUGAGCUAAUGGC-UCGUAAAUGGCUGGGCUAGCUGGAUUCGAACCAGCGCAUGACGGAGUC---AAAGUCCGUUGCCUUACCG .(((.((((((((.....)))))).((((..-......))))....)).-))).....((..((((..(((((.......)))))....((((((...---....))))))))))..)). ( -39.10) >Bhalo.0 558270 120 + 4202352/320-440 CUAACCUUUUGAUCCGUAGUCAAACGCUCUAUCCAAUUGAGCUACGGGCGCUUUAUACGAUGCGGUUGAGAGGACUCGAACCUCCACGGGGUCAACCCCCACUAGGCCCUCAACCUAGCG .............((((((((((.............)))).))))))((((..........))(((((((.((.......(((....((((....))))....))))))))))))..)). ( -34.63) >consensus UCGACCCUCUGAUUAAAAGUCAGAUGCUCUA_CCAACUGAGCUAAUGGC_UCGUAAAUGGCUGGGCUAGCUGGAUUCGAACCAGCGCAUGACGGAGUC___AAAGUCCGUUGCCUUACCG ....((.((((((.....)))))).((((.........))))....)).........(((..((((..(((((.......)))))....((((((.(......).))))))))))..))) (-23.20 = -23.77 + 0.57)

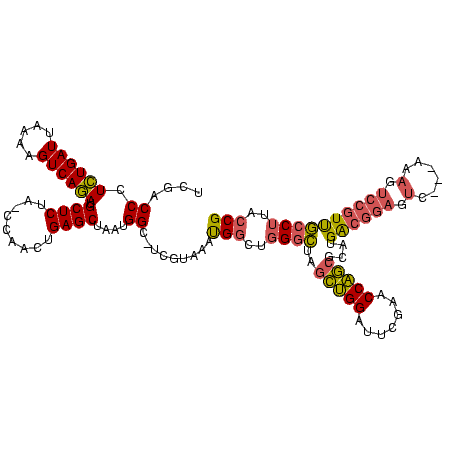
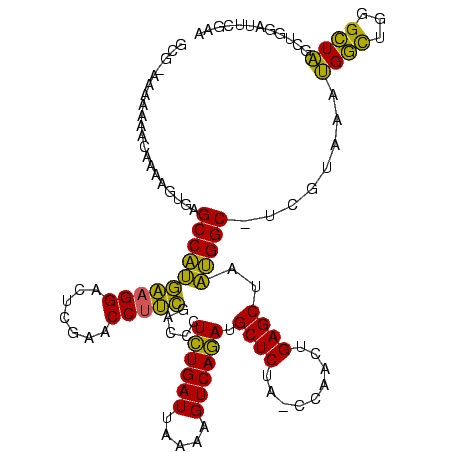
| Location | 245,259 – 245,377 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.79 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -24.66 |
| Energy contribution | -21.67 |
| Covariance contribution | -2.99 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 245259 118 + 5227293/360-480 GCGAAAAAAAACAAAAGUGAGCCAUGAAGGACUCGAACCUUCGACCCUCUGAUUAAAAGUCAGAUGCUCUA-CCAACUGAGCUAAUGGC-UCGUAAAUGGCUGGGCUAGCUGGAUUCGAA .((((.............((((((((((((.......))))).....((((((.....)))))).((((..-......))))..)))))-))......(((((...)))))...)))).. ( -34.10) >Bcere.0 278654 117 + 5224283/360-480 GCG-GAAAAAACAAAAGUGAGCCAUGAAGGACUCGAACCUUCGACCCUCUGAUUAAAAGUCAGAUGCUCUA-CCAACUGAGCUAAUGGC-UCGUAAAUGGCUGGGCUAGCUGGAUUCGAA .((-((............((((((((((((.......))))).....((((((.....)))))).((((..-......))))..)))))-))......(((((...)))))...)))).. ( -33.60) >Bhalo.0 558270 116 + 4202352/360-480 ACUCAAAACA----UGGCGCGCCCGAGAGGAGUCGAACCCCUAACCUUUUGAUCCGUAGUCAAACGCUCUAUCCAAUUGAGCUACGGGCGCUUUAUACGAUGCGGUUGAGAGGACUCGAA .(((((..((----(.(.(((((((...(((.(((((..........))))))))..........((((.........))))..)))))))......).)))...))))).......... ( -31.50) >consensus GCG_AAAAAAACAAAAGUGAGCCAUGAAGGACUCGAACCUUCGACCCUCUGAUUAAAAGUCAGAUGCUCUA_CCAACUGAGCUAAUGGC_UCGUAAAUGGCUGGGCUAGCUGGAUUCGAA ....................((((((((((.......))))).....((((((.....)))))).((((.........))))..)))))........((((...))))............ (-24.66 = -21.67 + -2.99)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:21:28 2006