
| Sequence ID | Bcere.0 |
|---|---|
| Location | 225,068 – 3,833,676 |
| Length | 3608608 |
| Max. P | 0.999997 |

| Location | 3,833,644 – 3,833,676 |
|---|---|
| Length | 32 |
| Sequences | 3 |
| Columns | 32 |
| Reading direction | forward |
| Mean pairwise identity | 85.42 |
| Mean single sequence MFE | -12.43 |
| Consensus MFE | -11.75 |
| Energy contribution | -12.53 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.16 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991621 |
| Prediction | RNA |
| WARNING | Sequence 1 too short. |
| WARNING | Sequence 2 too short. |
| WARNING | Sequence 3 too short. |
Download alignment: ClustalW | MAF
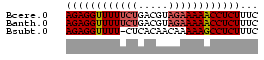
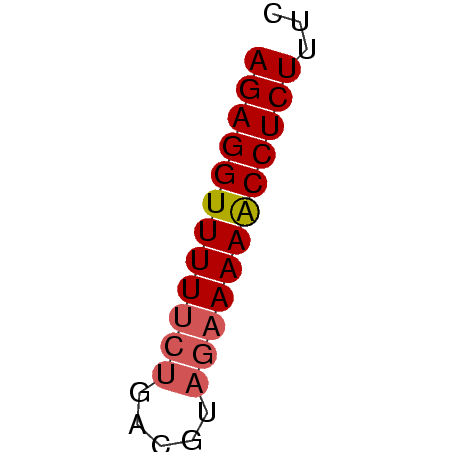
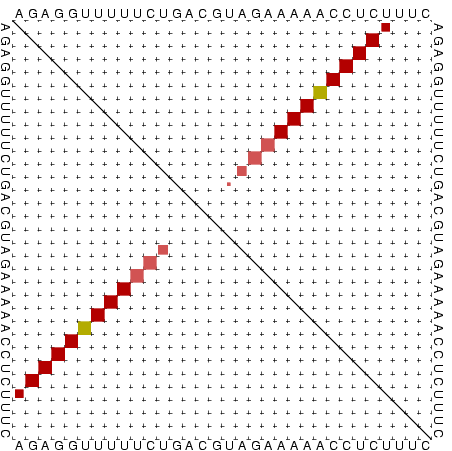
>Bcere.0 3833644 32 + 5224283 AGAGGUUUUUCUGACGUAGAAAAACCUCUUUC (((((((((((((...)))))))))))))... ( -14.60) >Banth.0 3890530 32 + 5227293 AGAGGUUUUUCUGACGUAGAAAAACCUCUUUC (((((((((((((...)))))))))))))... ( -14.60) >Bsubt.0 1384971 31 + 4214630 AGAGGUUUU-CUCACAACAAAAAGCCUCUUUC (((((((((-..........)))))))))... ( -8.10) >consensus AGAGGUUUUUCUGACGUAGAAAAACCUCUUUC ((((((((((((.....))))))))))))... (-11.75 = -12.53 + 0.78)
| Location | 3,833,644 – 3,833,676 |
|---|---|
| Length | 32 |
| Sequences | 3 |
| Columns | 32 |
| Reading direction | reverse |
| Mean pairwise identity | 85.42 |
| Mean single sequence MFE | -12.33 |
| Consensus MFE | -10.99 |
| Energy contribution | -11.77 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.85 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989885 |
| Prediction | RNA |
| WARNING | Sequence 1 too short. |
| WARNING | Sequence 2 too short. |
| WARNING | Sequence 3 too short. |
Download alignment: ClustalW | MAF
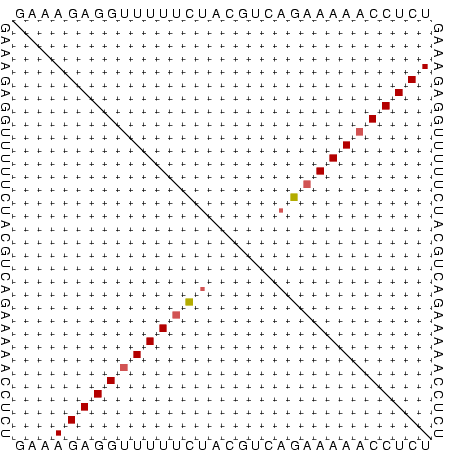
>Bcere.0 3833644 32 + 5224283 GAAAGAGGUUUUUCUACGUCAGAAAAACCUCU ...((((((((((((.....)))))))))))) ( -14.70) >Banth.0 3890530 32 + 5227293 GAAAGAGGUUUUUCUACGUCAGAAAAACCUCU ...((((((((((((.....)))))))))))) ( -14.70) >Bsubt.0 1384971 31 + 4214630 GAAAGAGGCUUUUUGUUGUGAG-AAAACCUCU ...(((((.(((((......))-))).))))) ( -7.60) >consensus GAAAGAGGUUUUUCUACGUCAGAAAAACCUCU ...((((((((((((.....)))))))))))) (-10.99 = -11.77 + 0.78)


| Location | 225,068 – 225,185 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.19 |
| Mean single sequence MFE | -35.07 |
| Consensus MFE | -20.09 |
| Energy contribution | -21.32 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.63 |
| Structure conservation index | 0.57 |
| SVM decision value | 6.08 |
| SVM RNA-class probability | 0.999996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 225068 117 + 5224283/0-120 AAAAAAUCCCUCUUUUCAUAUUGAGAAAGAGGGAUUUUUACGUACAA---GUUAUGCACCCGUAUAUAAACGAAGUCCCUCAUUCUAUCUUUCAAGCUAUAAAGCUUGCAGGAAGUGGCA .(((((((((((((((........))))))))))))))).(((....---..(((((....)))))...)))..(((.((.......((((.((((((....)))))).)))))).))). ( -33.31) >Banth.0 197002 117 + 5227293/0-120 AAAAAAUCCCUCUUUUCGUAUUGAGAAAGAGGGAUUUUUACGUACAA---GUUAUGCACCCGUAUAUAAACGAAGUCCCUCAUUCUAUCUUUCAAGCUAUAAAGCUUGCAGGAAGUGGCA .(((((((((((((((........))))))))))))))).(((....---..(((((....)))))...)))..(((.((.......((((.((((((....)))))).)))))).))). ( -33.31) >Bsubt.0 2024288 118 + 4214630/0-120 AAAAAAUCUCUUUCCGAAGAUGGAAAGAGAUUCAUAUAUAGGCGCGCUCUGUUACAGGCAGACGCCGGUAUAAACCUUGCUUUCCUAUCUUUCAAGCUA--GCGCUUGCUGGAAUUGGCA ....(((((((((((......)))))))))))..(((((.((((...(((((.....))))))))).))))).....((((......(((..(((((..--..)))))..)))...)))) ( -38.60) >consensus AAAAAAUCCCUCUUUUCAUAUUGAGAAAGAGGGAUUUUUACGUACAA___GUUAUGCACCCGUAUAUAAACGAAGUCCCUCAUUCUAUCUUUCAAGCUAUAAAGCUUGCAGGAAGUGGCA .(((((((((((((((........)))))))))))))))..((((................))))......................((((.(((((......))))).))))....... (-20.09 = -21.32 + 1.23)

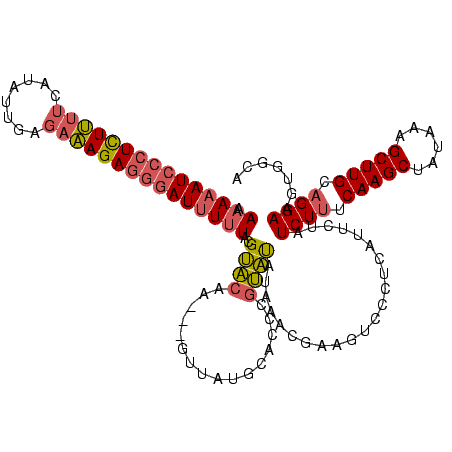

| Location | 225,068 – 225,185 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.19 |
| Mean single sequence MFE | -37.43 |
| Consensus MFE | -21.55 |
| Energy contribution | -22.23 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.79 |
| Structure conservation index | 0.58 |
| SVM decision value | 6.20 |
| SVM RNA-class probability | 0.999997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 225068 117 + 5224283/0-120 UGCCACUUCCUGCAAGCUUUAUAGCUUGAAAGAUAGAAUGAGGGACUUCGUUUAUAUACGGGUGCAUAAC---UUGUACGUAAAAAUCCCUCUUUCUCAAUAUGAAAAGAGGGAUUUUUU ..((...((...((((((....))))))...))..(((((((....)))))))......))(((((....---.)))))..((((((((((((((.((.....)))))))))))))))). ( -36.40) >Banth.0 197002 117 + 5227293/0-120 UGCCACUUCCUGCAAGCUUUAUAGCUUGAAAGAUAGAAUGAGGGACUUCGUUUAUAUACGGGUGCAUAAC---UUGUACGUAAAAAUCCCUCUUUCUCAAUACGAAAAGAGGGAUUUUUU ..((...((...((((((....))))))...))..(((((((....)))))))......))(((((....---.)))))..((((((((((((((.((.....)))))))))))))))). ( -36.10) >Bsubt.0 2024288 118 + 4214630/0-120 UGCCAAUUCCAGCAAGCGC--UAGCUUGAAAGAUAGGAAAGCAAGGUUUAUACCGGCGUCUGCCUGUAACAGAGCGCGCCUAUAUAUGAAUCUCUUUCCAUCUUCGGAAAGAGAUUUUUU (((...((((..(((((..--..))))).......)))).))).(((....)))(((((..(((((...))).))))))).......((((((((((((......))))))))))))... ( -39.80) >consensus UGCCACUUCCUGCAAGCUUUAUAGCUUGAAAGAUAGAAUGAGGGACUUCGUUUAUAUACGGGUGCAUAAC___UUGUACGUAAAAAUCCCUCUUUCUCAAUAUGAAAAGAGGGAUUUUUU .......((...(((((......)))))...))..(((((((....)))))))........(((((........)))))..((((((((((((((..........)))))))))))))). (-21.55 = -22.23 + 0.68)

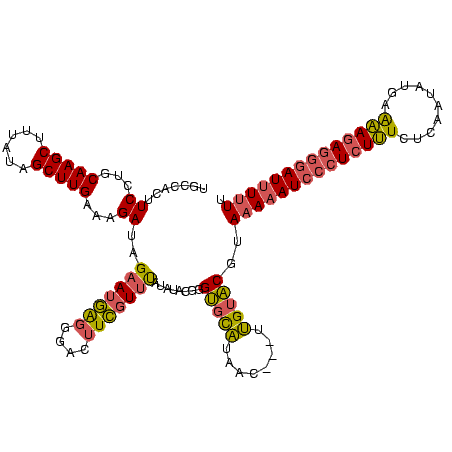
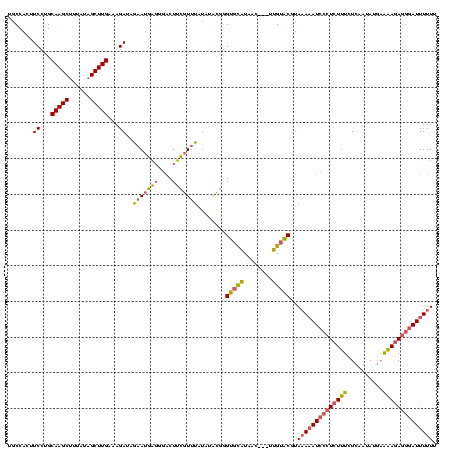
| Location | 225,068 – 225,185 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.98 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -16.39 |
| Energy contribution | -16.85 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 225068 117 + 5224283/40-160 CGUACAA---GUUAUGCACCCGUAUAUAAACGAAGUCCCUCAUUCUAUCUUUCAAGCUAUAAAGCUUGCAGGAAGUGGCACAGUAUUCAAAAAAUGAACCUGCUGCCGAGGUUUCAAAGG .......---..(((((....))))).....((((..((((......((((.((((((....)))))).))))...(((((((..((((.....)))).))).))))))))))))..... ( -29.90) >Banth.0 197002 116 + 5227293/40-160 CGUACAA---GUUAUGCACCCGUAUAUAAACGAAGUCCCUCAUUCUAUCUUUCAAGCUAUAAAGCUUGCAGGAAGUGGCACAGUAUUCAAAAA-UGAACCUGCUGCCGAGGUUUCAAAGG .......---..(((((....))))).....((((..((((......((((.((((((....)))))).))))...(((((((..((((....-)))).))).))))))))))))..... ( -29.90) >Bsubt.0 2024288 104 + 4214630/40-160 GGCGCGCUCUGUUACAGGCAGACGCCGGUAUAAACCUUGCUUUCCUAUCUUUCAAGCUA--GCGCUUGCUGGAAUUGGCACAAUAAU--------------GUUGCCGAGGUUUCAUAGG ((((...(((((.....)))))))))..((((((((((.........(((..(((((..--..)))))..)))...((((((....)--------------).))))))))))).))).. ( -32.20) >consensus CGUACAA___GUUAUGCACCCGUAUAUAAACGAAGUCCCUCAUUCUAUCUUUCAAGCUAUAAAGCUUGCAGGAAGUGGCACAGUAUUCAAAAA_UGAACCUGCUGCCGAGGUUUCAAAGG .((((................))))............((((......((((.(((((......))))).))))...(((((((................))).))))))))......... (-16.39 = -16.85 + 0.45)

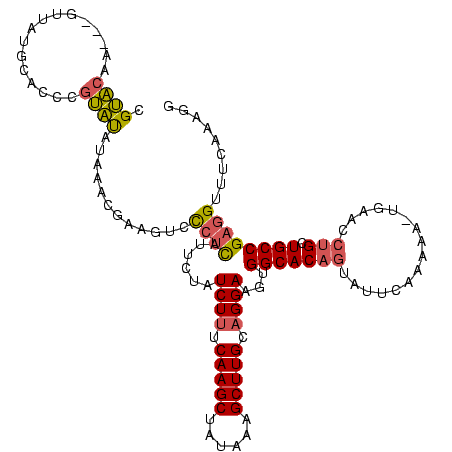

| Location | 225,068 – 225,185 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.98 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -21.43 |
| Energy contribution | -22.83 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.64 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 225068 117 + 5224283/40-160 CCUUUGAAACCUCGGCAGCAGGUUCAUUUUUUGAAUACUGUGCCACUUCCUGCAAGCUUUAUAGCUUGAAAGAUAGAAUGAGGGACUUCGUUUAUAUACGGGUGCAUAAC---UUGUACG .........((((((((.((((((((.....))))).))))))).((((...((((((....))))))...)).))...))))....((((......))))(((((....---.))))). ( -35.20) >Banth.0 197002 116 + 5227293/40-160 CCUUUGAAACCUCGGCAGCAGGUUCA-UUUUUGAAUACUGUGCCACUUCCUGCAAGCUUUAUAGCUUGAAAGAUAGAAUGAGGGACUUCGUUUAUAUACGGGUGCAUAAC---UUGUACG .........((((((((.((((((((-....))))).))))))).((((...((((((....))))))...)).))...))))....((((......))))(((((....---.))))). ( -35.20) >Bsubt.0 2024288 104 + 4214630/40-160 CCUAUGAAACCUCGGCAAC--------------AUUAUUGUGCCAAUUCCAGCAAGCGC--UAGCUUGAAAGAUAGGAAAGCAAGGUUUAUACCGGCGUCUGCCUGUAACAGAGCGCGCC (((((........((((.(--------------(....))))))........(((((..--..)))))....))))).......(((....)))(((((..(((((...))).))))))) ( -30.70) >consensus CCUUUGAAACCUCGGCAGCAGGUUCA_UUUUUGAAUACUGUGCCACUUCCUGCAAGCUUUAUAGCUUGAAAGAUAGAAUGAGGGACUUCGUUUAUAUACGGGUGCAUAAC___UUGUACG ....((((.((((((((.((((((((.....))))).)))))))...((...(((((......)))))...))......))))...))))...........(((((........))))). (-21.43 = -22.83 + 1.40)

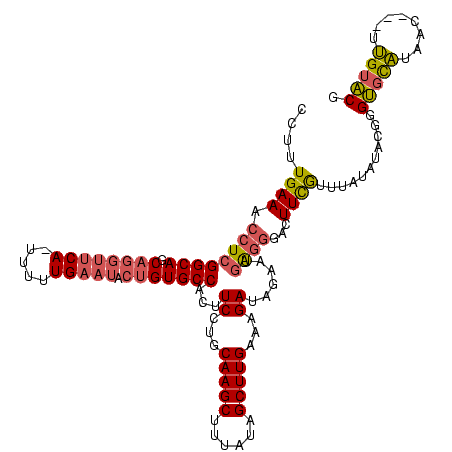
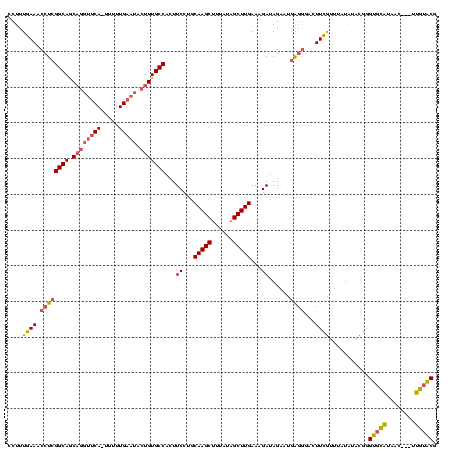
| Location | 225,068 – 225,183 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.23 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -24.33 |
| Energy contribution | -24.56 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 225068 115 + 5224283/79-199 UCAUUCUAUCUUUCAAGCUAUAAAGCUUGCAGGAAGUGGCACAGUAUUCAAAAAAUGAACCUGCUGCCGAGGUUUCAAAGGGCCAGUCCCUCCACCUCUCUUGAUACAA-----UGAGUA (((((.((((...((((((....)))))).((((...(((((((..((((.....)))).))).))))(((((.....((((.....))))..))))))))))))).))-----)))... ( -33.80) >Banth.0 197002 114 + 5227293/79-199 UCAUUCUAUCUUUCAAGCUAUAAAGCUUGCAGGAAGUGGCACAGUAUUCAAAAA-UGAACCUGCUGCCGAGGUUUCAAAGGGCCAGUCCCUCCACCUCUCUUGAUACAA-----UGAGUA (((((.((((...((((((....)))))).((((...(((((((..((((....-)))).))).))))(((((.....((((.....))))..))))))))))))).))-----)))... ( -33.80) >Bsubt.0 2024288 104 + 4214630/79-199 CUUUCCUAUCUUUCAAGCUA--GCGCUUGCUGGAAUUGGCACAAUAAU--------------GUUGCCGAGGUUUCAUAGGGCCAGUCCCUCCACCUCUCUGGAUAGAAAAUAUUGAGUA ((.(((..(((..(((((..--..)))))..)))...((((((....)--------------).))))(((((.....((((.....))))..)))))...))).))............. ( -27.60) >consensus UCAUUCUAUCUUUCAAGCUAUAAAGCUUGCAGGAAGUGGCACAGUAUUCAAAAA_UGAACCUGCUGCCGAGGUUUCAAAGGGCCAGUCCCUCCACCUCUCUUGAUACAA_____UGAGUA .............(((((......)))))(((((...(((((((................))).))))(((((.....((((.....))))..))))))))))................. (-24.33 = -24.56 + 0.22)

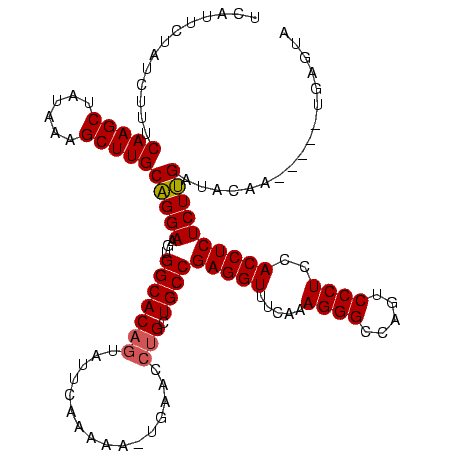
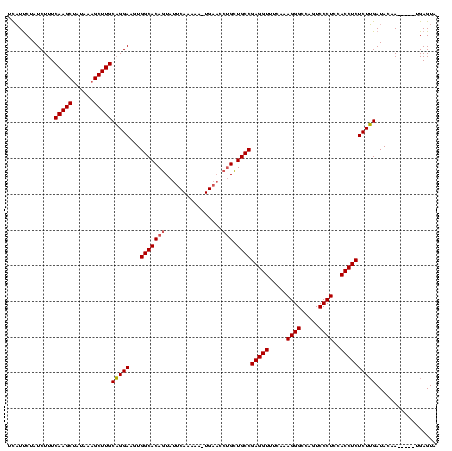
| Location | 225,068 – 225,183 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.23 |
| Mean single sequence MFE | -40.23 |
| Consensus MFE | -28.69 |
| Energy contribution | -30.63 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.97 |
| Structure conservation index | 0.71 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 225068 115 + 5224283/79-199 UACUCA-----UUGUAUCAAGAGAGGUGGAGGGACUGGCCCUUUGAAACCUCGGCAGCAGGUUCAUUUUUUGAAUACUGUGCCACUUCCUGCAAGCUUUAUAGCUUGAAAGAUAGAAUGA ...(((-----((.(((((((.(((((.(((((.....)))))....)))))((((.((((((((.....))))).))))))).)))....((((((....))))))...)))).))))) ( -43.00) >Banth.0 197002 114 + 5227293/79-199 UACUCA-----UUGUAUCAAGAGAGGUGGAGGGACUGGCCCUUUGAAACCUCGGCAGCAGGUUCA-UUUUUGAAUACUGUGCCACUUCCUGCAAGCUUUAUAGCUUGAAAGAUAGAAUGA ...(((-----((.(((((((.(((((.(((((.....)))))....)))))((((.((((((((-....))))).))))))).)))....((((((....))))))...)))).))))) ( -43.00) >Bsubt.0 2024288 104 + 4214630/79-199 UACUCAAUAUUUUCUAUCCAGAGAGGUGGAGGGACUGGCCCUAUGAAACCUCGGCAAC--------------AUUAUUGUGCCAAUUCCAGCAAGCGC--UAGCUUGAAAGAUAGGAAAG .........(((((((((....(((((..((((.....)))).....)))))((((.(--------------(....))))))........(((((..--..)))))...))))))))). ( -34.70) >consensus UACUCA_____UUGUAUCAAGAGAGGUGGAGGGACUGGCCCUUUGAAACCUCGGCAGCAGGUUCA_UUUUUGAAUACUGUGCCACUUCCUGCAAGCUUUAUAGCUUGAAAGAUAGAAUGA ...........((.((((....(((((.(((((.....)))))....)))))((((.((((((((.....))))).)))))))........(((((......)))))...)))).))... (-28.69 = -30.63 + 1.95)

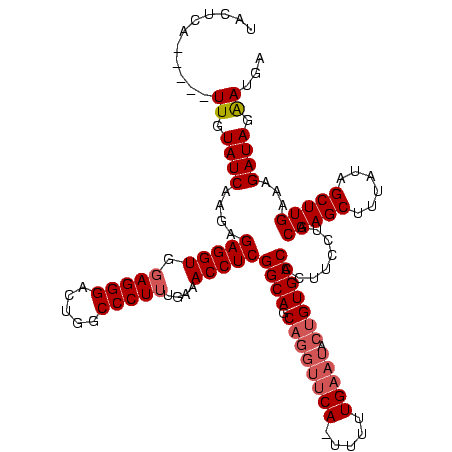
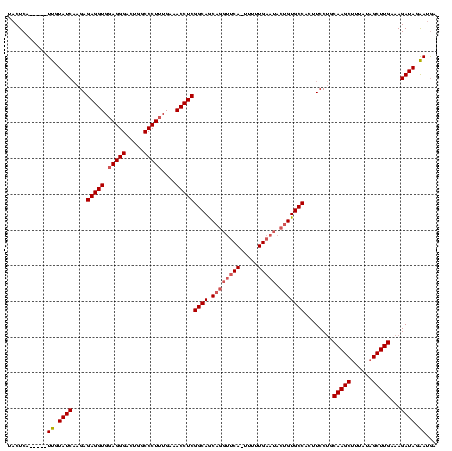
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:21:22 2006