
| Sequence ID | Bcere.0 |
|---|---|
| Location | 2,730,666 – 3,425,135 |
| Length | 694469 |
| Max. P | 0.999955 |

| Location | 3,425,079 – 3,425,135 |
|---|---|
| Length | 56 |
| Sequences | 3 |
| Columns | 56 |
| Reading direction | forward |
| Mean pairwise identity | 66.67 |
| Mean single sequence MFE | -9.87 |
| Consensus MFE | -5.63 |
| Energy contribution | -6.97 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.57 |
| SVM decision value | 4.84 |
| SVM RNA-class probability | 0.999955 |
| Prediction | RNA |
| WARNING | Sequence 1: Base composition out of range. |
Download alignment: ClustalW | MAF
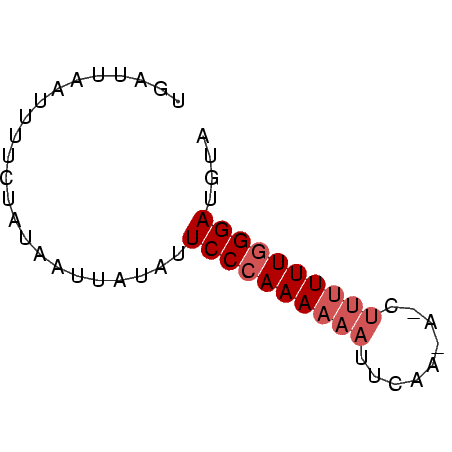
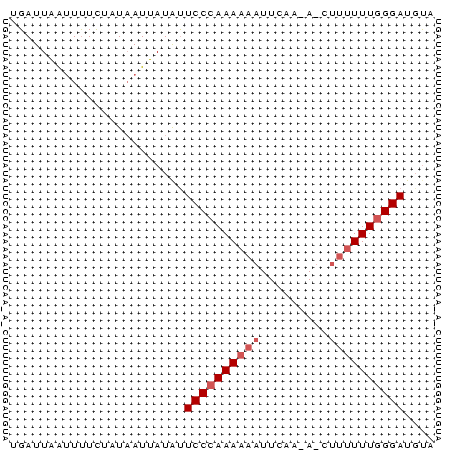
>Bcere.0 3425079 56 + 5224283 UGAUUAAUUUUCUAUAAUUAUGUUCCCAAAAAAUUCAAAACCUUUUUUGGGAUCUA ((((((........))))))...((((((((((.........)))))))))).... ( -10.20) >Banth.0 3409724 56 + 5227293 UGAUUAAUUCUCUAUAAUUAUACUCCCAAAAAAUUCAAGAUCUUUUUUGGGAUGCC ((((((........))))))...((((((((((.........)))))))))).... ( -10.10) >Bsubt.0 559647 46 + 4214630 AGAUUUGCUUUUCAUCAUAAUAUUCCGAAAUA----------UCUUUUGGGAUGUA ...................(((((((((((..----------..))))))))))). ( -9.30) >consensus UGAUUAAUUUUCUAUAAUUAUAUUCCCAAAAAAUUCAA_A_CUUUUUUGGGAUGUA .......................((((((((((.........)))))))))).... ( -5.63 = -6.97 + 1.33)

| Location | 3,425,079 – 3,425,135 |
|---|---|
| Length | 56 |
| Sequences | 3 |
| Columns | 56 |
| Reading direction | reverse |
| Mean pairwise identity | 66.67 |
| Mean single sequence MFE | -8.53 |
| Consensus MFE | -7.05 |
| Energy contribution | -7.83 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.83 |
| SVM decision value | 4.00 |
| SVM RNA-class probability | 0.999750 |
| Prediction | RNA |
| WARNING | Sequence 1: Base composition out of range. |
Download alignment: ClustalW | MAF
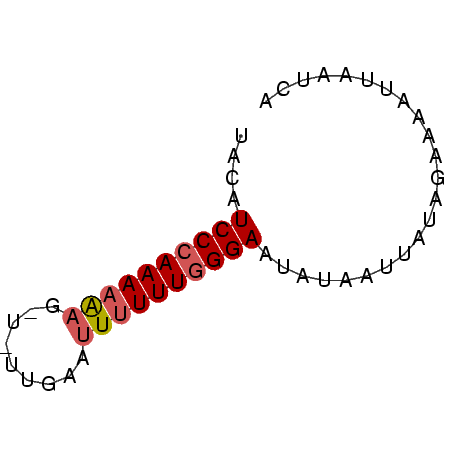
>Bcere.0 3425079 56 + 5224283 UAGAUCCCAAAAAAGGUUUUGAAUUUUUUGGGAACAUAAUUAUAGAAAAUUAAUCA ..(.((((((((((.........)))))))))).)..................... ( -11.10) >Banth.0 3409724 56 + 5227293 GGCAUCCCAAAAAAGAUCUUGAAUUUUUUGGGAGUAUAAUUAUAGAGAAUUAAUCA .((.((((((((((.........))))))))))))..................... ( -11.70) >Bsubt.0 559647 46 + 4214630 UACAUCCCAAAAGA----------UAUUUCGGAAUAUUAUGAUGAAAAGCAAAUCU ..((((......((----------((((....))))))..))))............ ( -2.80) >consensus UACAUCCCAAAAAAG_U_UUGAAUUUUUUGGGAAUAUAAUUAUAGAAAAUUAAUCA ....((((((((((.........))))))))))....................... ( -7.05 = -7.83 + 0.78)
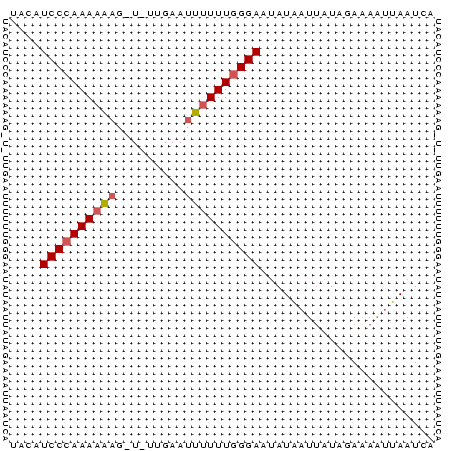
| Location | 2,730,666 – 2,730,784 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -16.10 |
| Energy contribution | -17.10 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 2730666 118 - 5224283/0-120 AUCCCUUUCAUUAA-GGGAGAAUAAAAAAAUCCUCCCAAAUAAAUUUGGCAGGAUUAGUCGUUAAAAUAUUUAUACCCAAAUAAGCUAUUGCGCAACAUAAAUAGCAAUACCGU-UGUAU .((((((.....))-)))).........((((((.(((((....))))).))))))............................((((((..........))))))........-..... ( -24.50) >Banth.0 2806177 117 - 5227293/0-120 AUCCCUUUCAUUAA-GGGAGAAUAAAAAA-UCCUCCCAAAUAAAUUUGGCAGGAUUAGUCGUUAAAAUAUUUAUACCCAAAUAAACUAUUGCGCAACAUAAAUAGCAAUACCGU-UGUAU .((((((.....))-))))........((-((((.(((((....))))).))))))........................((((..((((((............))))))...)-))).. ( -24.40) >Bsubt.0 605927 114 + 4214630/0-120 AUCCCUCGCUUUAAAGGGAGAAUAAAAAAAUCCUCCCAAAUGUGUU-GGAAGGAUUAGCCGU--GCCUGUGCAUGAUCAAAUAAGCUAUUUUU---UAUAAAUAGCAGUGUCGUAUAUAU .(((((........)))))((.......((((((.((((.....))-)).))))))...(((--((....))))).))......(((((((..---...))))))).............. ( -28.50) >consensus AUCCCUUUCAUUAA_GGGAGAAUAAAAAAAUCCUCCCAAAUAAAUUUGGCAGGAUUAGUCGUUAAAAUAUUUAUACCCAAAUAAGCUAUUGCGCAACAUAAAUAGCAAUACCGU_UGUAU .((((..........)))).........((((((.(((((....))))).))))))............................((((((..........)))))).............. (-16.10 = -17.10 + 1.00)

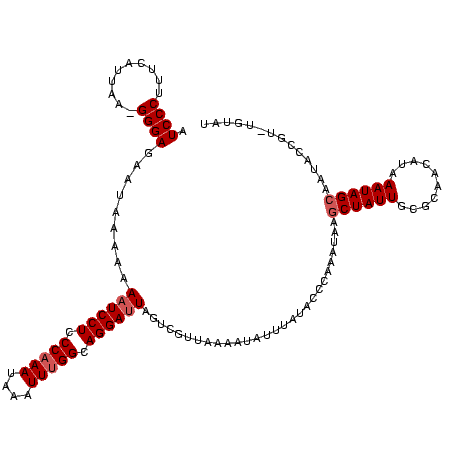
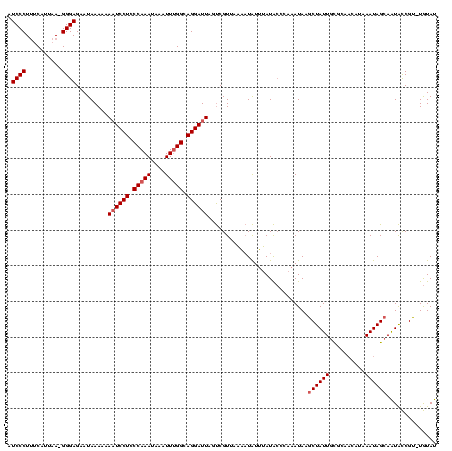
| Location | 2,730,666 – 2,730,784 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -18.74 |
| Energy contribution | -18.63 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 2730666 118 - 5224283/0-120 AUACA-ACGGUAUUGCUAUUUAUGUUGCGCAAUAGCUUAUUUGGGUAUAAAUAUUUUAACGACUAAUCCUGCCAAAUUUAUUUGGGAGGAUUUUUUUAUUCUCCC-UUAAUGAAAGGGAU ((((.-..((((..((((((..........)))))).))))...))))................((((((.(((((....))))).)))))).........((((-((.....)))))). ( -26.40) >Banth.0 2806177 117 - 5227293/0-120 AUACA-ACGGUAUUGCUAUUUAUGUUGCGCAAUAGUUUAUUUGGGUAUAAAUAUUUUAACGACUAAUCCUGCCAAAUUUAUUUGGGAGGA-UUUUUUAUUCUCCC-UUAAUGAAAGGGAU .....-.((.((((((............))))))((((((......)))))).......))...((((((.(((((....))))).))))-))........((((-((.....)))))). ( -26.30) >Bsubt.0 605927 114 + 4214630/0-120 AUAUAUACGACACUGCUAUUUAUA---AAAAAUAGCUUAUUUGAUCAUGCACAGGC--ACGGCUAAUCCUUCC-AACACAUUUGGGAGGAUUUUUUUAUUCUCCCUUUAAAGCGAGGGAU .......((.....(((((((...---..)))))))...........(((....))--)))...(((((((((-((.....))))))))))).........((((((......)))))). ( -29.20) >consensus AUACA_ACGGUAUUGCUAUUUAUGUUGCGCAAUAGCUUAUUUGGGUAUAAAUAUUUUAACGACUAAUCCUGCCAAAUUUAUUUGGGAGGAUUUUUUUAUUCUCCC_UUAAUGAAAGGGAU .......(((....((((((..........))))))....)))......................(((((......((((((.((((((((......))))))))...)))))).))))) (-18.74 = -18.63 + -0.10)

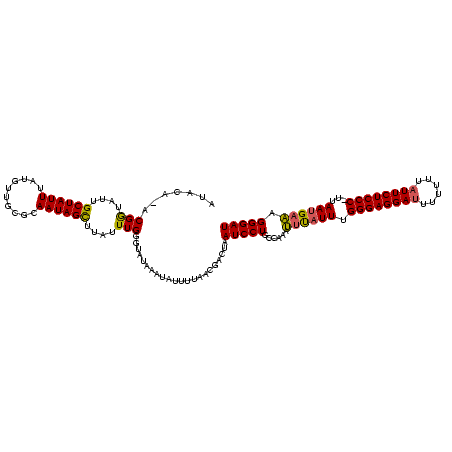
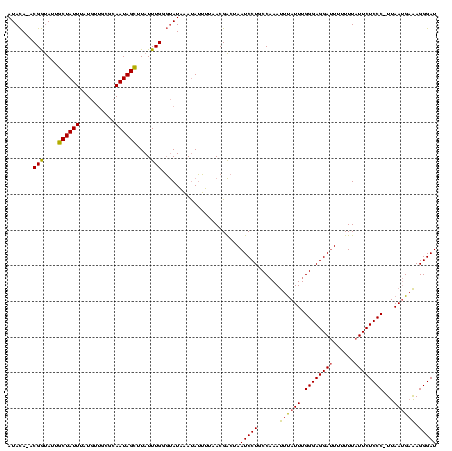
| Location | 2,730,666 – 2,730,785 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -18.99 |
| Energy contribution | -18.00 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 2730666 119 - 5224283/40-160 UAAAUUUGGCAGGAUUAGUCGUUAAAAUAUUUAUACCCAAAUAAGCUAUUGCGCAACAUAAAUAGCAAUACCGU-UGUAUGGAAAUUGGGCAGACUAAUCCUAUUUUUUAGAAAUUGAAA ..........((((((((((..(((.....)))..(((((....((((((..........))))))....((((-...))))...)))))..)))))))))).................. ( -26.40) >Banth.0 2806177 119 - 5227293/40-160 UAAAUUUGGCAGGAUUAGUCGUUAAAAUAUUUAUACCCAAAUAAACUAUUGCGCAACAUAAAUAGCAAUACCGU-UGUAUGGAAAUUGGGCAGACUAAUCCUAUUUUUUAGAAAUUGAAA ..........((((((((((..(((.....)))..(((((......((((((............))))))((((-...))))...)))))..)))))))))).................. ( -25.20) >Bsubt.0 605927 112 + 4214630/40-160 UGUGUU-GGAAGGAUUAGCCGU--GCCUGUGCAUGAUCAAAUAAGCUAUUUUU---UAUAAAUAGCAGUGUCGUAUAUAUAGAAAAUGGGCAGACUAAUCCUAUUUUUU--GAUUUGAAA ((..(.-((..((.....))..--.)).)..))...((((((..(((((((..---...))))))).............(((((((((((.(......)))))))))))--))))))).. ( -25.60) >consensus UAAAUUUGGCAGGAUUAGUCGUUAAAAUAUUUAUACCCAAAUAAGCUAUUGCGCAACAUAAAUAGCAAUACCGU_UGUAUGGAAAUUGGGCAGACUAAUCCUAUUUUUUAGAAAUUGAAA ..........((((((((((....((....))...((((.....((((((..........)))))).((((.....))))......))))..)))))))))).................. (-18.99 = -18.00 + -0.99)

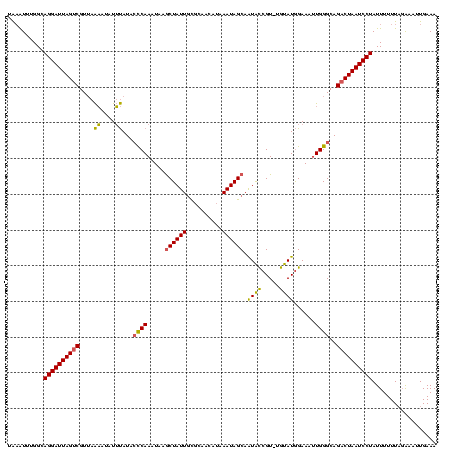
| Location | 2,730,666 – 2,730,785 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -19.22 |
| Energy contribution | -18.57 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.74 |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.998669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 2730666 119 - 5224283/40-160 UUUCAAUUUCUAAAAAAUAGGAUUAGUCUGCCCAAUUUCCAUACA-ACGGUAUUGCUAUUUAUGUUGCGCAAUAGCUUAUUUGGGUAUAAAUAUUUUAACGACUAAUCCUGCCAAAUUUA .................((((((((((((((((((.....((((.-...)))).((((((..........))))))....)))))))(((.....)))..)))))))))))......... ( -26.40) >Banth.0 2806177 119 - 5227293/40-160 UUUCAAUUUCUAAAAAAUAGGAUUAGUCUGCCCAAUUUCCAUACA-ACGGUAUUGCUAUUUAUGUUGCGCAAUAGUUUAUUUGGGUAUAAAUAUUUUAACGACUAAUCCUGCCAAAUUUA .................((((((((((((((((((...((.....-..))((((((............))))))......)))))))(((.....)))..)))))))))))......... ( -26.10) >Bsubt.0 605927 112 + 4214630/40-160 UUUCAAAUC--AAAAAAUAGGAUUAGUCUGCCCAUUUUCUAUAUAUACGACACUGCUAUUUAUA---AAAAAUAGCUUAUUUGAUCAUGCACAGGC--ACGGCUAAUCCUUCC-AACACA .........--.......(((((((((((((((((..((...(((.........(((((((...---..))))))).)))..))..)))....)))--).))))))))))...-...... ( -25.00) >consensus UUUCAAUUUCUAAAAAAUAGGAUUAGUCUGCCCAAUUUCCAUACA_ACGGUAUUGCUAUUUAUGUUGCGCAAUAGCUUAUUUGGGUAUAAAUAUUUUAACGACUAAUCCUGCCAAAUUUA .................(((((((((((((((((.......(((.....)))..((((((..........)))))).....)))))).((....))....)))))))))))......... (-19.22 = -18.57 + -0.66)

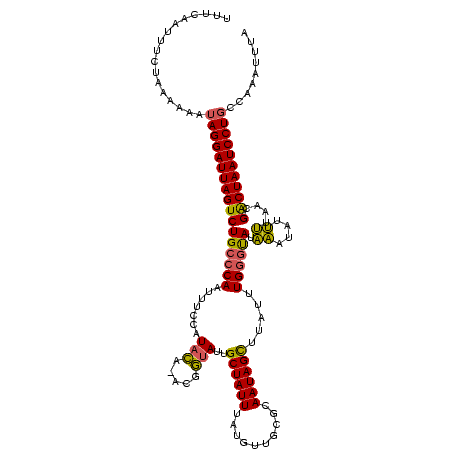
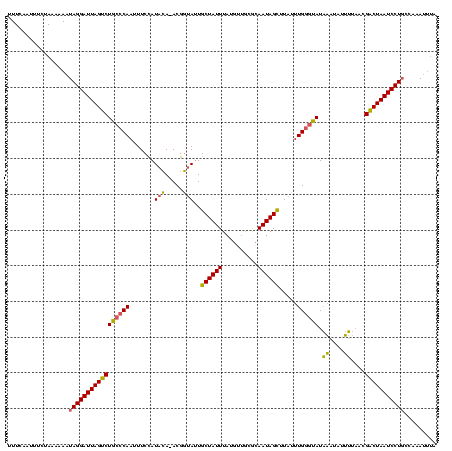
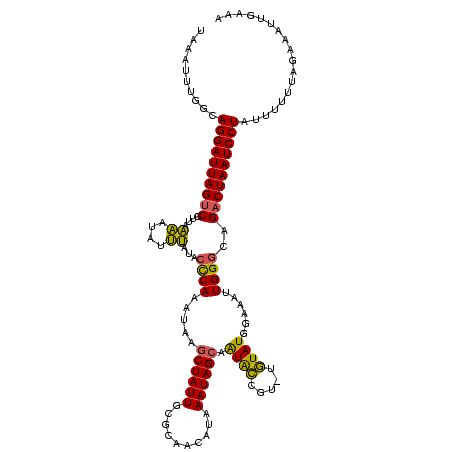
| Location | 2,730,666 – 2,730,785 |
|---|---|
| Length | 119 |
| Sequences | 2 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -25.15 |
| Energy contribution | -25.15 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 2730666 119 - 5224283/80-200 AUAAGCUAUUGCGCAACAUAAAUAGCAAUACCGUUGUAUGGAAAUUGGGCAGACUAAUCCUAUUUUUUAGAAAUUGAAAUGUAUUGAAUUUCAACAUUUAAAAAUAAGAUUAGUUAAUC ....((((((((............))))).((((...))))......))).((((((((.(((((((.(((..(((((((.......)))))))..)))))))))).)))))))).... ( -25.70) >Banth.0 2806177 119 - 5227293/80-200 AUAAACUAUUGCGCAACAUAAAUAGCAAUACCGUUGUAUGGAAAUUGGGCAGACUAAUCCUAUUUUUUAGAAAUUGAAAUUUAUUGAAUUUCAACAUUUAAAAAUAAGAUUAGUUAAUC .........(((.(((..((.(((((......))))).))....))).)))((((((((.(((((((.(((..(((((((((...)))))))))..)))))))))).)))))))).... ( -26.20) >consensus AUAAACUAUUGCGCAACAUAAAUAGCAAUACCGUUGUAUGGAAAUUGGGCAGACUAAUCCUAUUUUUUAGAAAUUGAAAUGUAUUGAAUUUCAACAUUUAAAAAUAAGAUUAGUUAAUC .........(((.(((..((.(((((......))))).))....))).)))((((((((.(((((((.(((..(((((((.......)))))))..)))))))))).)))))))).... (-25.15 = -25.15 + 0.00)

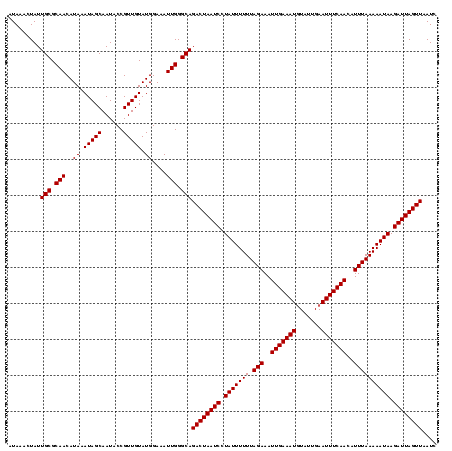
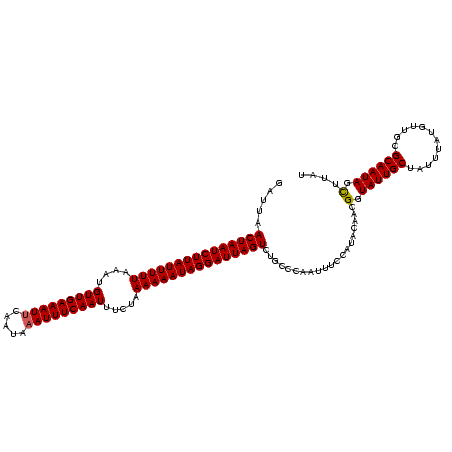
| Location | 2,730,666 – 2,730,785 |
|---|---|
| Length | 119 |
| Sequences | 2 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -24.30 |
| Energy contribution | -24.55 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 2730666 119 - 5224283/80-200 GAUUAACUAAUCUUAUUUUUAAAUGUUGAAAUUCAAUACAUUUCAAUUUCUAAAAAAUAGGAUUAGUCUGCCCAAUUUCCAUACAACGGUAUUGCUAUUUAUGUUGCGCAAUAGCUUAU .....(((((((((((((((....((((((((.......)))))))).....)))))))))))))))....................(.((((((............)))))).).... ( -24.90) >Banth.0 2806177 119 - 5227293/80-200 GAUUAACUAAUCUUAUUUUUAAAUGUUGAAAUUCAAUAAAUUUCAAUUUCUAAAAAAUAGGAUUAGUCUGCCCAAUUUCCAUACAACGGUAUUGCUAUUUAUGUUGCGCAAUAGUUUAU ((((((((((((((((((((....(((((((((.....))))))))).....))))))))))))))).(((.((((....((((((.....))).)))....)))).))).)))))... ( -25.50) >consensus GAUUAACUAAUCUUAUUUUUAAAUGUUGAAAUUCAAUAAAUUUCAAUUUCUAAAAAAUAGGAUUAGUCUGCCCAAUUUCCAUACAACGGUAUUGCUAUUUAUGUUGCGCAAUAGCUUAU .....(((((((((((((((....(((((((((.....))))))))).....)))))))))))))))....................(.((((((............)))))).).... (-24.30 = -24.55 + 0.25)

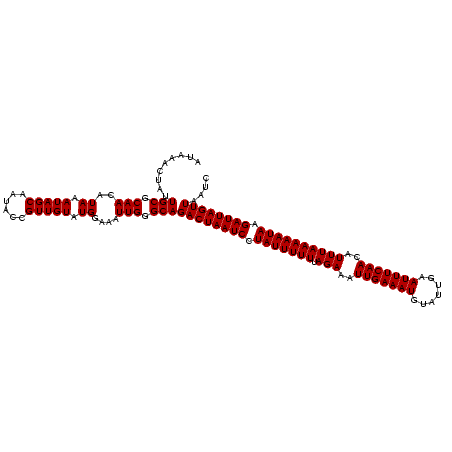
| Location | 2,730,666 – 2,730,786 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -19.07 |
| Energy contribution | -19.73 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 2730666 120 - 5224283/120-240 GGAAAUUGGGCAGACUAAUCCUAUUUUUUAGAAAUUGAAAUGUAUUGAAUUUCAACAUUUAAAAAUAAGAUUAGUUAAUCAUCGUCCACCCAUCAUUCCUUUCCGUCGUUAAUAACCAUA ((((..((((..((((((((.(((((((.(((..(((((((.......)))))))..)))))))))).))))))))............))))...))))..................... ( -23.84) >Banth.0 2806177 120 - 5227293/120-240 GGAAAUUGGGCAGACUAAUCCUAUUUUUUAGAAAUUGAAAUUUAUUGAAUUUCAACAUUUAAAAAUAAGAUUAGUUAAUCAUCGUCCACCCAUCAUUCCUUUCCGUCAUUAAUAACCAUA ((((..((((..((((((((.(((((((.(((..(((((((((...)))))))))..)))))))))).))))))))............))))...))))..................... ( -24.34) >Bsubt.0 605927 115 + 4214630/120-240 AGAAAAUGGGCAGACUAAUCCUAUUUUUU--GAUUUGAAAU--AUUGA-UUUCAAACGGUAAAAAUAAGAUUAGUUAAUCAUUGUCCACCCAUCAUCCCUUCUUGUUUUGAUUACAAACA ......((((((((((((((.(((((((.--..((((((((--....)-)))))))....))))))).))))))))......))))))..............((((.......))))... ( -23.90) >consensus GGAAAUUGGGCAGACUAAUCCUAUUUUUUAGAAAUUGAAAU_UAUUGAAUUUCAACAUUUAAAAAUAAGAUUAGUUAAUCAUCGUCCACCCAUCAUUCCUUUCCGUCAUUAAUAACCAUA (((...(((...((((((((.(((((((......(((((((.......))))))).....))))))).))))))))..)))...)))................................. (-19.07 = -19.73 + 0.67)


| Location | 2,730,666 – 2,730,786 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -33.03 |
| Consensus MFE | -29.92 |
| Energy contribution | -29.37 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.09 |
| Mean z-score | -4.21 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 2730666 120 - 5224283/120-240 UAUGGUUAUUAACGACGGAAAGGAAUGAUGGGUGGACGAUGAUUAACUAAUCUUAUUUUUAAAUGUUGAAAUUCAAUACAUUUCAAUUUCUAAAAAAUAGGAUUAGUCUGCCCAAUUUCC ....(((......))).....((((...((((..((((........)(((((((((((((....((((((((.......)))))))).....))))))))))))))))..))))..)))) ( -31.00) >Banth.0 2806177 120 - 5227293/120-240 UAUGGUUAUUAAUGACGGAAAGGAAUGAUGGGUGGACGAUGAUUAACUAAUCUUAUUUUUAAAUGUUGAAAUUCAAUAAAUUUCAAUUUCUAAAAAAUAGGAUUAGUCUGCCCAAUUUCC ....((((....)))).....((((...((((..((((........)(((((((((((((....(((((((((.....))))))))).....))))))))))))))))..))))..)))) ( -33.10) >Bsubt.0 605927 115 + 4214630/120-240 UGUUUGUAAUCAAAACAAGAAGGGAUGAUGGGUGGACAAUGAUUAACUAAUCUUAUUUUUACCGUUUGAAA-UCAAU--AUUUCAAAUC--AAAAAAUAGGAUUAGUCUGCCCAUUUUCU (((((.......))))).....(((.((((((..(((..........(((((((((((((...((((((((-(....--))))))))).--.))))))))))))))))..)))))).))) ( -35.00) >consensus UAUGGUUAUUAAAGACGGAAAGGAAUGAUGGGUGGACGAUGAUUAACUAAUCUUAUUUUUAAAUGUUGAAAUUCAAUA_AUUUCAAUUUCUAAAAAAUAGGAUUAGUCUGCCCAAUUUCC .....................((((...((((..(((..........(((((((((((((....((((((((.......)))))))).....))))))))))))))))..))))..)))) (-29.92 = -29.37 + -0.55)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:21:16 2006