| Sequence ID | Bcere.0 |
|---|---|
| Location | 2,236,464 – 2,236,579 |
| Length | 115 |
| Max. P | 0.999014 |

| Location | 2,236,464 – 2,236,579 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.63 |
| Mean single sequence MFE | -22.53 |
| Consensus MFE | -13.82 |
| Energy contribution | -13.60 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 2236464 115 - 5224283/200-320 AUCUAGGAGAUCAGAAAUAUACUUUUCUUUUUAAUAGGAGUAAUACAUACAUU-----CUUUUAUAAAAUGGGAGCUCUUCAAAAAACUUUAUUGAAGUUAGUUGAAGGGAAACCGACAA ...(((((((..(((((......)))))...........(((.....)))..)-----)))))).....(((...((((((((..((((((...))))))..))))))))...))).... ( -23.30) >Banth.0 2295039 120 - 5227293/200-320 AUCUAGGAGAUCAGAGAUAUACUGUUAUUUUUAAUAGGAUUAAUACAUACAUUAUUAUCCUCUAUAAAAUGGGAGCUCUUCAAAAAACUUUAUUGAAGUUAGUUGAAGUGAAACCGACAA ((((..(....)..))))....(((((((((....(((((.((((.......)))))))))....)))))((...(.((((((..((((((...))))))..)))))).)...)))))). ( -23.00) >Bsubt.0 204931 109 + 4214630/200-320 AU------AAUCAGGACUAUA---UUAAAGGAAAUGAAAACAUAGCAUAAAGUA--AAGACCAAUAAUAAGGUAGCUCUUCAAAAAACUUCGCUGAAGUUAGUUGAAGUGAAGCCGACAA ..------..((.((.((.((---((....(..(((....)))..)....))))--.)).))........(((..(.((((((..((((((...))))))..)))))).)..)))))... ( -21.30) >consensus AUCUAGGAGAUCAGAAAUAUACU_UUAUUUUUAAUAGGAAUAAUACAUACAUUA__A_CAUCUAUAAAAUGGGAGCUCUUCAAAAAACUUUAUUGAAGUUAGUUGAAGUGAAACCGACAA ......................................................................((...(.((((((..((((((...))))))..)))))).)...))..... (-13.82 = -13.60 + -0.22)

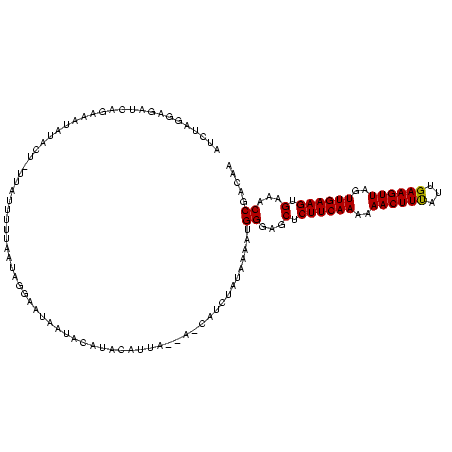
| Location | 2,236,464 – 2,236,579 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -26.95 |
| Energy contribution | -26.73 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -4.33 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 2236464 115 - 5224283/240-360 UAAUACAUACAUU-----CUUUUAUAAAAUGGGAGCUCUUCAAAAAACUUUAUUGAAGUUAGUUGAAGGGAAACCGACAAUCUCCAUAAUUCAAUGAUGAUAUAAAAUCAUUGAAUUAUG .............-----...........(((...((((((((..((((((...))))))..))))))))...)))........((((((((((((((........)))))))))))))) ( -32.80) >Banth.0 2295039 120 - 5227293/240-360 UAAUACAUACAUUAUUAUCCUCUAUAAAAUGGGAGCUCUUCAAAAAACUUUAUUGAAGUUAGUUGAAGUGAAACCGACAAUCUCCAUAAUUCAAUGAUGAUAUAAAAUCAUUGAAUUAUG .............................(((...(.((((((..((((((...))))))..)))))).)...)))........((((((((((((((........)))))))))))))) ( -29.50) >Bsubt.0 204931 118 + 4214630/240-360 CAUAGCAUAAAGUA--AAGACCAAUAAUAAGGUAGCUCUUCAAAAAACUUCGCUGAAGUUAGUUGAAGUGAAGCCGACAAUUUUCAGGAUUCAAUGAUCUAACAUAAUCAUUGAAAUUUU ..............--....((........(((..(.((((((..((((((...))))))..)))))).)..)))((......)).)).(((((((((........)))))))))..... ( -26.60) >consensus UAAUACAUACAUUA__A_CAUCUAUAAAAUGGGAGCUCUUCAAAAAACUUUAUUGAAGUUAGUUGAAGUGAAACCGACAAUCUCCAUAAUUCAAUGAUGAUAUAAAAUCAUUGAAUUAUG ..............................((...(.((((((..((((((...))))))..)))))).)...)).........((((((((((((((........)))))))))))))) (-26.95 = -26.73 + -0.22)

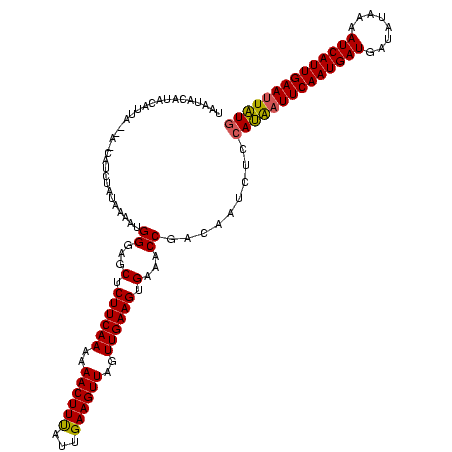
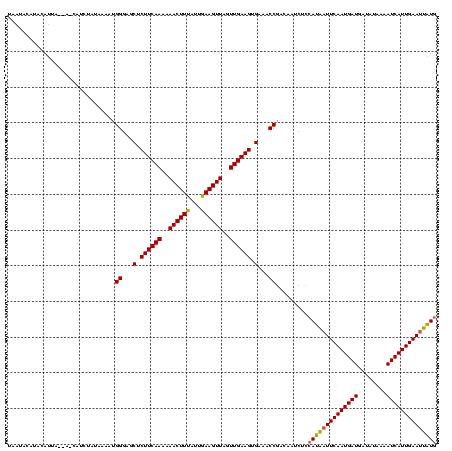
| Location | 2,236,464 – 2,236,579 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -22.51 |
| Energy contribution | -24.07 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 2236464 115 - 5224283/240-360 CAUAAUUCAAUGAUUUUAUAUCAUCAUUGAAUUAUGGAGAUUGUCGGUUUCCCUUCAACUAACUUCAAUAAAGUUUUUUGAAGAGCUCCCAUUUUAUAAAAG-----AAUGUAUGUAUUA .(((((((((((((........)))))))))))))((((((.....))))))((((((..(((((.....)))))..)))))).((...((((((.....))-----))))...)).... ( -28.70) >Banth.0 2295039 120 - 5227293/240-360 CAUAAUUCAAUGAUUUUAUAUCAUCAUUGAAUUAUGGAGAUUGUCGGUUUCACUUCAACUAACUUCAAUAAAGUUUUUUGAAGAGCUCCCAUUUUAUAGAGGAUAAUAAUGUAUGUAUUA ((((((((((((((........))))))))))))))...((((((.......((((((..(((((.....)))))..))))))..(((..........)))))))))............. ( -29.40) >Bsubt.0 204931 118 + 4214630/240-360 AAAAUUUCAAUGAUUAUGUUAGAUCAUUGAAUCCUGAAAAUUGUCGGCUUCACUUCAACUAACUUCAGCGAAGUUUUUUGAAGAGCUACCUUAUUAUUGGUCUU--UACUUUAUGCUAUG .....((((((((((......))))))))))..............((((...((((((..((((((...))))))..))))))))))(((........)))...--.............. ( -25.40) >consensus CAUAAUUCAAUGAUUUUAUAUCAUCAUUGAAUUAUGGAGAUUGUCGGUUUCACUUCAACUAACUUCAAUAAAGUUUUUUGAAGAGCUCCCAUUUUAUAGAAG_U__UAAUGUAUGUAUUA ((((((((((((((........)))))))))))))).(((.((..((((...((((((..(((((.....)))))..))))))))))..)).)))......................... (-22.51 = -24.07 + 1.56)

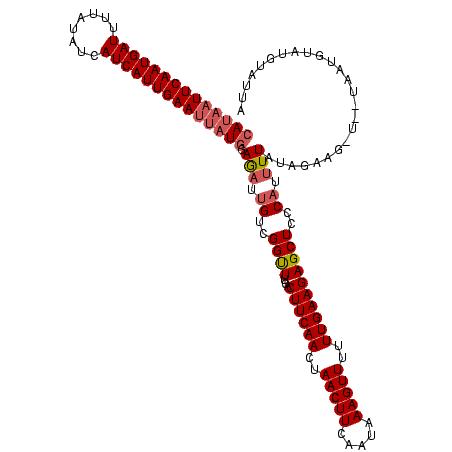
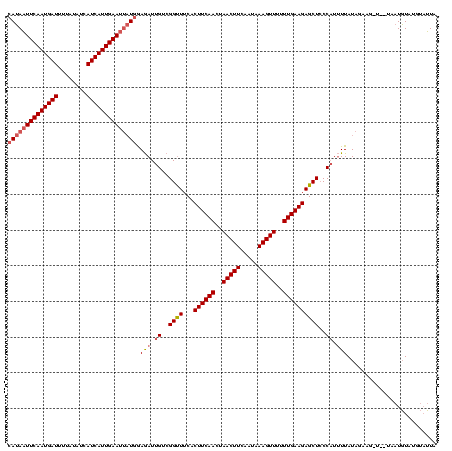
| Location | 2,236,464 – 2,236,579 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -31.00 |
| Energy contribution | -30.57 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -5.43 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 2236464 115 - 5224283/242-362 AUACAUACAUU-----CUUUUAUAAAAUGGGAGCUCUUCAAAAAACUUUAUUGAAGUUAGUUGAAGGGAAACCGACAAUCUCCAUAAUUCAAUGAUGAUAUAAAAUCAUUGAAUUAUGGA ...........-----...........(((...((((((((..((((((...))))))..))))))))...)))......((((((((((((((((........)))))))))))))))) ( -37.80) >Banth.0 2295039 120 - 5227293/242-362 AUACAUACAUUAUUAUCCUCUAUAAAAUGGGAGCUCUUCAAAAAACUUUAUUGAAGUUAGUUGAAGUGAAACCGACAAUCUCCAUAAUUCAAUGAUGAUAUAAAAUCAUUGAAUUAUGGA ...........................(((...(.((((((..((((((...))))))..)))))).)...)))......((((((((((((((((........)))))))))))))))) ( -34.50) >Bsubt.0 204931 118 + 4214630/242-362 UAGCAUAAAGUA--AAGACCAAUAAUAAGGUAGCUCUUCAAAAAACUUCGCUGAAGUUAGUUGAAGUGAAGCCGACAAUUUUCAGGAUUCAAUGAUCUAACAUAAUCAUUGAAAUUUUGA ............--..............(((..(.((((((..((((((...))))))..)))))).)..)))........(((((((((((((((........))))))).)))))))) ( -28.50) >consensus AUACAUACAUUA__A_CAUCUAUAAAAUGGGAGCUCUUCAAAAAACUUUAUUGAAGUUAGUUGAAGUGAAACCGACAAUCUCCAUAAUUCAAUGAUGAUAUAAAAUCAUUGAAUUAUGGA ............................((...(.((((((..((((((...))))))..)))))).)...)).......((((((((((((((((........)))))))))))))))) (-31.00 = -30.57 + -0.44)

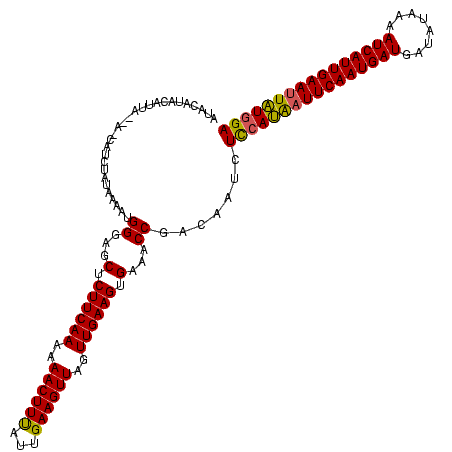
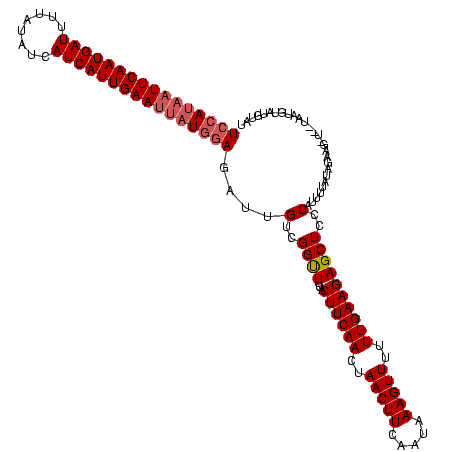
| Location | 2,236,464 – 2,236,579 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -25.39 |
| Energy contribution | -26.83 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.26 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 2236464 115 - 5224283/242-362 UCCAUAAUUCAAUGAUUUUAUAUCAUCAUUGAAUUAUGGAGAUUGUCGGUUUCCCUUCAACUAACUUCAAUAAAGUUUUUUGAAGAGCUCCCAUUUUAUAAAAG-----AAUGUAUGUAU ((((((((((((((((........)))))))))))))))).......((...(.((((((..(((((.....)))))..)))))).)..))((((((.....))-----))))....... ( -31.80) >Banth.0 2295039 120 - 5227293/242-362 UCCAUAAUUCAAUGAUUUUAUAUCAUCAUUGAAUUAUGGAGAUUGUCGGUUUCACUUCAACUAACUUCAAUAAAGUUUUUUGAAGAGCUCCCAUUUUAUAGAGGAUAAUAAUGUAUGUAU ((((((((((((((((........)))))))))))))))).((((((.......((((((..(((((.....)))))..))))))..(((..........)))))))))........... ( -34.10) >Bsubt.0 204931 118 + 4214630/242-362 UCAAAAUUUCAAUGAUUAUGUUAGAUCAUUGAAUCCUGAAAAUUGUCGGCUUCACUUCAACUAACUUCAGCGAAGUUUUUUGAAGAGCUACCUUAUUAUUGGUCUU--UACUUUAUGCUA (((....((((((((((......))))))))))...)))........((((...((((((..((((((...))))))..))))))))))(((........)))...--............ ( -26.50) >consensus UCCAUAAUUCAAUGAUUUUAUAUCAUCAUUGAAUUAUGGAGAUUGUCGGUUUCACUUCAACUAACUUCAAUAAAGUUUUUUGAAGAGCUCCCAUUUUAUAGAAG_U__UAAUGUAUGUAU ((((((((((((((((........))))))))))))))))....(..((((...((((((..(((((.....)))))..))))))))))..)............................ (-25.39 = -26.83 + 1.45)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:21:08 2006