| Sequence ID | Bsubt.0 |
|---|---|
| Location | 3,128,197 – 3,128,305 |
| Length | 108 |
| Max. P | 0.993502 |

| Location | 3,128,197 – 3,128,305 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 53.96 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -12.78 |
| Energy contribution | -11.65 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 3128197 108 + 4214630/0-120 AAAACCUUUUCC------AUCGAGGAAAGGGUUUGGUCU--U-UGUGCCUUUCAC-UCUUAUCGCUCAAGGA-AUCAUACAA-CCUUGCAACAGGUUAGCACCUUGGUUGUCUCACUCAG .(((((((((((------.....)))))))))))(((((--(-((.((..(....-....)..)).))))).-)))..((((-((.(((.........)))....))))))......... ( -28.40) >Blich.0 3080386 112 + 4222334/0-120 AAAACCUUUUCCUAUUC-GGUGAGGAAAGGGUCUGGUAU--UGCGUGCCUUUCAC-UCUUAUCGCUCAAGGU-AUUGAACA--CCUUGCAUCAGGUUAGCACCGUAGUUGUCUCAGA-AA ....(((((((((....-....)))))))))(((((.((--(((((((..((((.-...((((......)))-).)))).(--(((......))))..))).))))))....)))))-.. ( -29.30) >Bhalo.0 3427305 93 + 4202352/0-120 AAAACCCUUUUCCAUU---AUGAGGAAAAGGGUUAAU-U--AACAUACCUUUCGCCUCUUAUCUUUCAAGGC-AACG------CCUUGC---AGGUUAGCACCUU------UUCA----- ..(((((((((((...---....)))))))))))...-.--............((.....((((..((((((-...)------))))).---))))..)).....------....----- ( -26.10) >Bclau.0 3008296 86 + 4303871/0-120 AAGCCCUUUUCC-------AUGAGGAAAAGGGCUGUC----CAUUUACCUUUCGCCUCUUAUCUUCCAGAAC-GCCGAGCA--UUCUGC---AGGUUAGCACC----------------- .(((((((((((-------....)))))))))))...----............((.....((((..(((((.-((...)).--))))).---))))..))...----------------- ( -27.10) >Banth.0 4553493 112 + 5227293/0-120 AAAACCUUUCCUACUUUAUAUAGAGGAAAGGUUUAAUUUGCUACUUGCAUACUGCUUUUUGCCCUUCGCUCUUAUCGUUCGAGACCUGU---AGCCUCGCACAG-----GUUUUAGCACC .(((((((((((.((......)))))))))))))....((((....((.....((.....)).....))...........(((((((((---.((...))))))-----))))))))).. ( -32.70) >Bcere.0 4541111 112 + 5224283/0-120 AAAACCUUUCCUACAUAAUAGAGAGGAAAGGUUUAAUUUGCUACUUGCAUACUGCUUUUUGCCCUUCGCUCUUAUCGUUCGAGACCUGU---AGCCUCGCACAG-----GUUUUAGCACC .(((((((((((.(........))))))))))))....((((....((.....((.....)).....))...........(((((((((---.((...))))))-----))))))))).. ( -31.10) >consensus AAAACCUUUUCC__UU___AUGAGGAAAAGGUUUAAU_U__UACUUGCCUUUCGC_UCUUAUCCUUCAAGGU_AUCGUACA__CCCUGC___AGGUUAGCACCG_____GUUUCAG____ .(((((((((((...........)))))))))))...................................................................................... (-12.78 = -11.65 + -1.13)
| Location | 3,128,197 – 3,128,305 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 53.96 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -13.22 |
| Energy contribution | -10.92 |
| Covariance contribution | -2.30 |
| Combinations/Pair | 1.65 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 3128197 108 + 4214630/0-120 CUGAGUGAGACAACCAAGGUGCUAACCUGUUGCAAGG-UUGUAUGAU-UCCUUGAGCGAUAAGA-GUGAAAGGCACA-A--AGACCAAACCCUUUCCUCGAU------GGAAAAGGUUUU ((..(((..((((((.((((....)))).......))-)))).....-.((((..((.......-))..))))))).-.--))...(((((.(((((.....------))))).))))). ( -27.90) >Blich.0 3080386 112 + 4222334/0-120 UU-UCUGAGACAACUACGGUGCUAACCUGAUGCAAGG--UGUUCAAU-ACCUUGAGCGAUAAGA-GUGAAAGGCACGCA--AUACCAGACCCUUUCCUCACC-GAAUAGGAAAAGGUUUU ..-...............(((((.((((......)))--)((((((.-...)))))).......-......)))))...--.....(((((.((((((....-....)))))).))))). ( -26.60) >Bhalo.0 3427305 93 + 4202352/0-120 -----UGAA------AAGGUGCUAACCU---GCAAGG------CGUU-GCCUUGAAAGAUAAGAGGCGAAAGGUAUGUU--A-AUUAACCCUUUUCCUCAU---AAUGGAAAAGGGUUUU -----....------((.(((((..((.---....))------..((-(((((.........)))))))..))))).))--.-...(((((((((((....---...))))))))))).. ( -28.80) >Bclau.0 3008296 86 + 4303871/0-120 -----------------GGUGCUAACCU---GCAGAA--UGCUCGGC-GUUCUGGAAGAUAAGAGGCGAAAGGUAAAUG----GACAGCCCUUUUCCUCAU-------GGAAAAGGGCUU -----------------(((....))).---.(((((--(((...))-))))))............(....).......----...(((((((((((....-------))))))))))). ( -32.00) >Banth.0 4553493 112 + 5227293/0-120 GGUGCUAAAAC-----CUGUGCGAGGCU---ACAGGUCUCGAACGAUAAGAGCGAAGGGCAAAAAGCAGUAUGCAAGUAGCAAAUUAAACCUUUCCUCUAUAUAAAGUAGGAAAGGUUUU ..(((((....-----.(((.((((((.---....)))))).)))......(((....((.....))....)))...)))))....(((((((((((((......)).))))))))))). ( -34.00) >Bcere.0 4541111 112 + 5224283/0-120 GGUGCUAAAAC-----CUGUGCGAGGCU---ACAGGUCUCGAACGAUAAGAGCGAAGGGCAAAAAGCAGUAUGCAAGUAGCAAAUUAAACCUUUCCUCUCUAUUAUGUAGGAAAGGUUUU ..(((((....-----.(((.((((((.---....)))))).)))......(((....((.....))....)))...)))))....((((((((((((........).))))))))))). ( -33.20) >consensus ____CUGAAAC_____CGGUGCUAACCU___GCAAGG__UGAACGAU_ACCUUGAAAGAUAAGA_GCGAAAGGCAAGUA__A_AUCAAACCUUUUCCUCAU___AA__GGAAAAGGUUUU ...................((((..(((......)))......((.......)).................))))...........(((((((((((...........))))))))))). (-13.22 = -10.92 + -2.30)

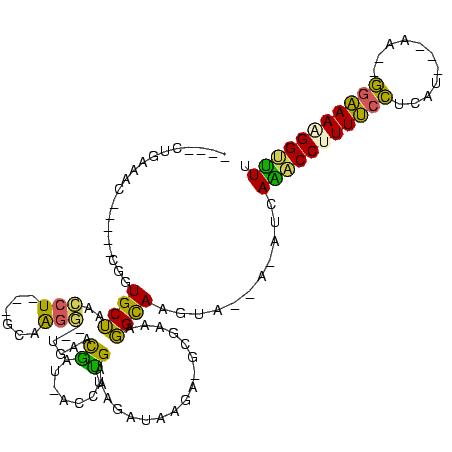
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:15:04 2006