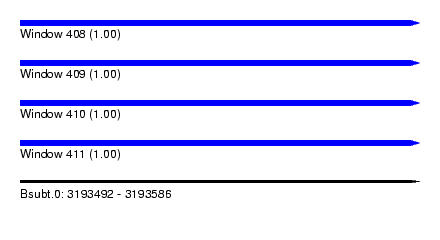
| Sequence ID | Bsubt.0 |
|---|---|
| Location | 3,193,492 – 3,193,586 |
| Length | 94 |
| Max. P | 0.999445 |

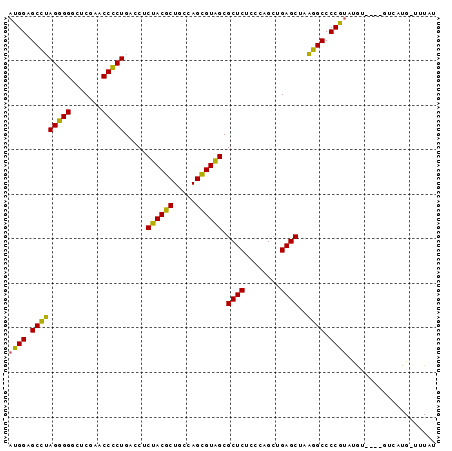
| Location | 3,193,492 – 3,193,586 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -33.07 |
| Energy contribution | -30.97 |
| Covariance contribution | -2.10 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.13 |
| Structure conservation index | 1.06 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 3193492 94 + 4214630 AUGGAGCCAAGGGGGCUCGAACCCCUGACCUCUACGCUGCCAGCGUAGCGCUCUCCCAGCUGAGCUAUGGCCCCGUAUGUA-ACGUCAUG-UUUAU ((((.(((((((((.......))))).....((((((.....)))))).((((........))))..)))).)))).....-........-..... ( -33.20) >Bclau.0 3031149 90 + 4303871 AUGGAGCCUAGGGGGCUCGAACCCCUGACCUCAACGCUGCCAGCGUUGCGCUCUCCCAGCUGAGCUAAGGCCCCACACG-----GUUAUG-UAUGU .(((.(((((((((.......))))).....((((((.....)))))).((((........))))..)))).)))....-----......-..... ( -32.00) >Blich.0 3143410 96 + 4222334 AUGGAGCAUAGCGGGCUCGAACCGCUGACCUCUACACUGCCAGUGUAGCGCUCUCCCAGCUGAGCUAAUGCCCCGUAUAUUUGAGACAUGAUUUAU ((((.(((((((((.......))))).....((((((.....)))))).((((........))))..)))).)))).................... ( -28.50) >consensus AUGGAGCCUAGGGGGCUCGAACCCCUGACCUCUACGCUGCCAGCGUAGCGCUCUCCCAGCUGAGCUAAGGCCCCGUAUGU____GUCAUG_UUUAU ((((.(((((((((.......))))).....((((((.....)))))).((((........))))..)))).)))).................... (-33.07 = -30.97 + -2.10)

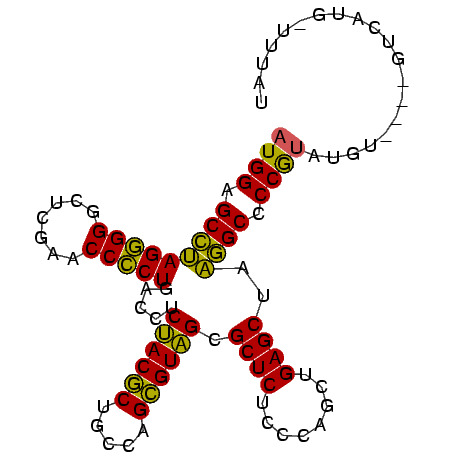
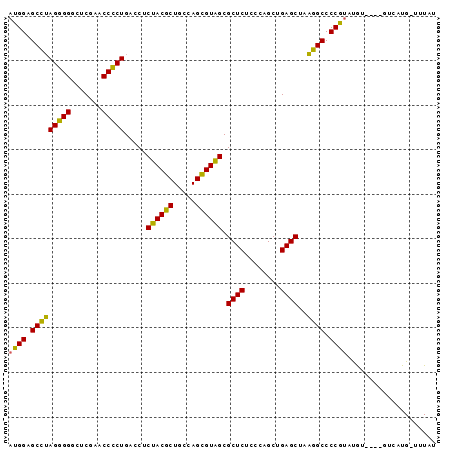
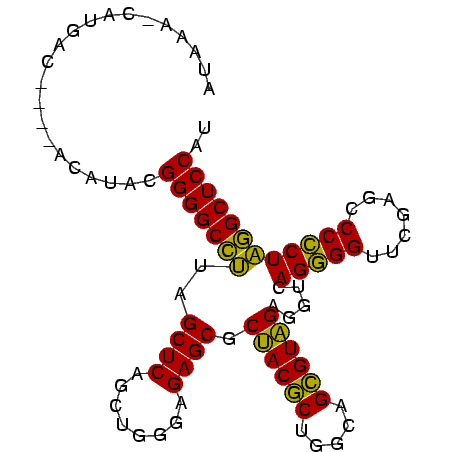
| Location | 3,193,492 – 3,193,586 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -35.05 |
| Energy contribution | -32.83 |
| Covariance contribution | -2.22 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.43 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 3193492 94 + 4214630 AUAAA-CAUGACGU-UACAUACGGGGCCAUAGCUCAGCUGGGAGAGCGCUACGCUGGCAGCGUAGAGGUCAGGGGUUCGAGCCCCCUUGGCUCCAU .....-........-.......(((((((..((((........)))).((((((.....))))))......(((((....)))))..))))))).. ( -35.90) >Bclau.0 3031149 90 + 4303871 ACAUA-CAUAAC-----CGUGUGGGGCCUUAGCUCAGCUGGGAGAGCGCAACGCUGGCAGCGUUGAGGUCAGGGGUUCGAGCCCCCUAGGCUCCAU .....-......-----...(((((((((.(((((....))).((.(.((((((.....)))))).).)).(((((....)))))))))))))))) ( -36.20) >Blich.0 3143410 96 + 4222334 AUAAAUCAUGUCUCAAAUAUACGGGGCAUUAGCUCAGCUGGGAGAGCGCUACACUGGCAGUGUAGAGGUCAGCGGUUCGAGCCCGCUAUGCUCCAU ......................(((((((.(((...(((.(((..((.((((((.....))))))......))..))).)))..)))))))))).. ( -32.10) >consensus AUAAA_CAUGAC____ACAUACGGGGCCUUAGCUCAGCUGGGAGAGCGCUACGCUGGCAGCGUAGAGGUCAGGGGUUCGAGCCCCCUAGGCUCCAU ......................(((((((..((((........)))).((((((.....)))))).....(((((.......)))))))))))).. (-35.05 = -32.83 + -2.22)

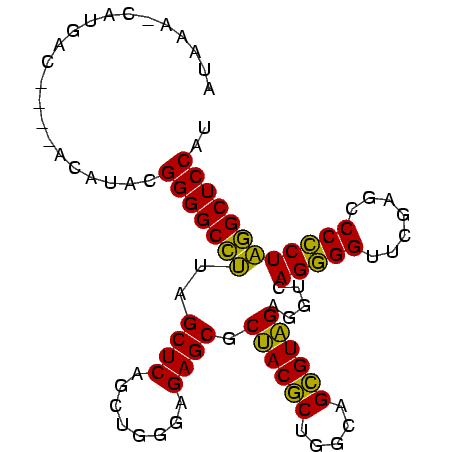
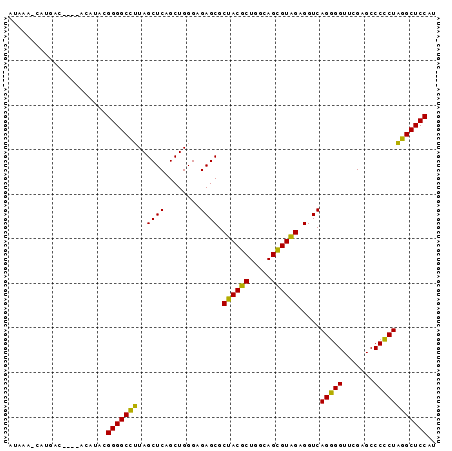
| Location | 3,193,492 – 3,193,586 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -33.07 |
| Energy contribution | -30.97 |
| Covariance contribution | -2.10 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.13 |
| Structure conservation index | 1.06 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 3193492 94 + 4214630/0-96 AUGGAGCCAAGGGGGCUCGAACCCCUGACCUCUACGCUGCCAGCGUAGCGCUCUCCCAGCUGAGCUAUGGCCCCGUAUGUA-ACGUCAUG-UUUAU ((((.(((((((((.......))))).....((((((.....)))))).((((........))))..)))).)))).....-........-..... ( -33.20) >Bclau.0 3031149 90 + 4303871/0-96 AUGGAGCCUAGGGGGCUCGAACCCCUGACCUCAACGCUGCCAGCGUUGCGCUCUCCCAGCUGAGCUAAGGCCCCACACG-----GUUAUG-UAUGU .(((.(((((((((.......))))).....((((((.....)))))).((((........))))..)))).)))....-----......-..... ( -32.00) >Blich.0 3143410 96 + 4222334/0-96 AUGGAGCAUAGCGGGCUCGAACCGCUGACCUCUACACUGCCAGUGUAGCGCUCUCCCAGCUGAGCUAAUGCCCCGUAUAUUUGAGACAUGAUUUAU ((((.(((((((((.......))))).....((((((.....)))))).((((........))))..)))).)))).................... ( -28.50) >consensus AUGGAGCCUAGGGGGCUCGAACCCCUGACCUCUACGCUGCCAGCGUAGCGCUCUCCCAGCUGAGCUAAGGCCCCGUAUGU____GUCAUG_UUUAU ((((.(((((((((.......))))).....((((((.....)))))).((((........))))..)))).)))).................... (-33.07 = -30.97 + -2.10)

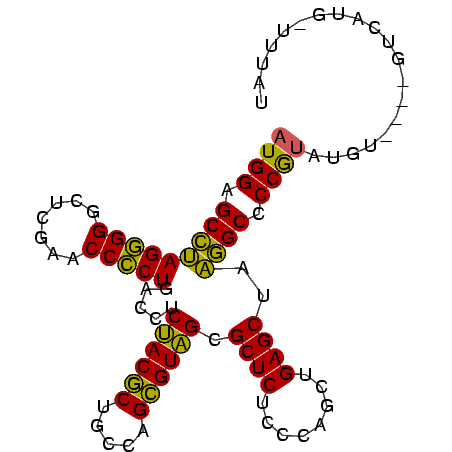
| Location | 3,193,492 – 3,193,586 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -35.05 |
| Energy contribution | -32.83 |
| Covariance contribution | -2.22 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.43 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 3193492 94 + 4214630/0-96 AUAAA-CAUGACGU-UACAUACGGGGCCAUAGCUCAGCUGGGAGAGCGCUACGCUGGCAGCGUAGAGGUCAGGGGUUCGAGCCCCCUUGGCUCCAU .....-........-.......(((((((..((((........)))).((((((.....))))))......(((((....)))))..))))))).. ( -35.90) >Bclau.0 3031149 90 + 4303871/0-96 ACAUA-CAUAAC-----CGUGUGGGGCCUUAGCUCAGCUGGGAGAGCGCAACGCUGGCAGCGUUGAGGUCAGGGGUUCGAGCCCCCUAGGCUCCAU .....-......-----...(((((((((.(((((....))).((.(.((((((.....)))))).).)).(((((....)))))))))))))))) ( -36.20) >Blich.0 3143410 96 + 4222334/0-96 AUAAAUCAUGUCUCAAAUAUACGGGGCAUUAGCUCAGCUGGGAGAGCGCUACACUGGCAGUGUAGAGGUCAGCGGUUCGAGCCCGCUAUGCUCCAU ......................(((((((.(((...(((.(((..((.((((((.....))))))......))..))).)))..)))))))))).. ( -32.10) >consensus AUAAA_CAUGAC____ACAUACGGGGCCUUAGCUCAGCUGGGAGAGCGCUACGCUGGCAGCGUAGAGGUCAGGGGUUCGAGCCCCCUAGGCUCCAU ......................(((((((..((((........)))).((((((.....)))))).....(((((.......)))))))))))).. (-35.05 = -32.83 + -2.22)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:21:00 2006