
| Sequence ID | Bsubt.0 |
|---|---|
| Location | 559,018 – 559,096 |
| Length | 78 |
| Max. P | 0.996733 |

| Location | 559,018 – 559,096 |
|---|---|
| Length | 78 |
| Sequences | 3 |
| Columns | 81 |
| Reading direction | forward |
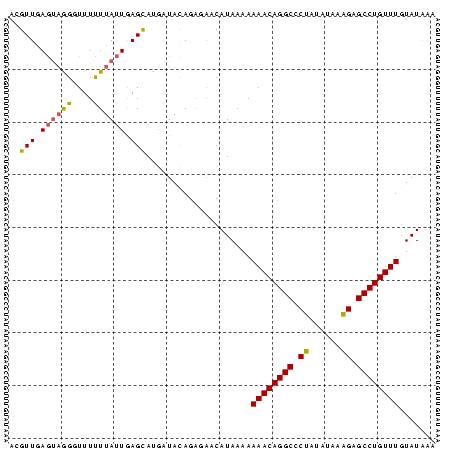
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -18.37 |
| Consensus MFE | -15.27 |
| Energy contribution | -15.17 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.84 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
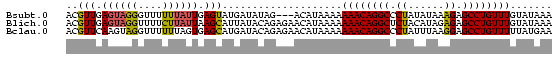
>Bsubt.0 559018 78 + 4214630 UUUAUACAAACAGGCUCUUUAUAUAGGGCCUGUUUUUUUAUGU---CUAUAUCAUACUCAAUAAAAAACCCUACUCAACGU (((((..(((((((((((......)))))))))))...((((.---......))))....)))))................ ( -16.60) >Blich.0 578017 81 + 4222334 UUUAUACAAACAGGCUCUCUAUGUAGAGCCUGUUUUUUUAUGUUCUCUGUAUAAUGCUUAAUAAGAAAACCUACUCAACGU .(((((((((((((((((......)))))))))).............)))))))........................... ( -18.21) >Bclau.0 902341 81 + 4303871 UUCAUAAAAACAGGCUCCUUAAAUAGGGCCUGUUUUUUUAUGUUCUCUGUAUCAUGCUCACUAAAAAACCUACUUGAACGU .....((((((((((.(((.....)))))))))))))..((((((...(((...................)))..)))))) ( -20.31) >consensus UUUAUACAAACAGGCUCUUUAUAUAGGGCCUGUUUUUUUAUGUUCUCUGUAUCAUGCUCAAUAAAAAACCCUACUCAACGU ..((((.(((((((((((......)))))))))))...))))....................................... (-15.27 = -15.17 + -0.11)

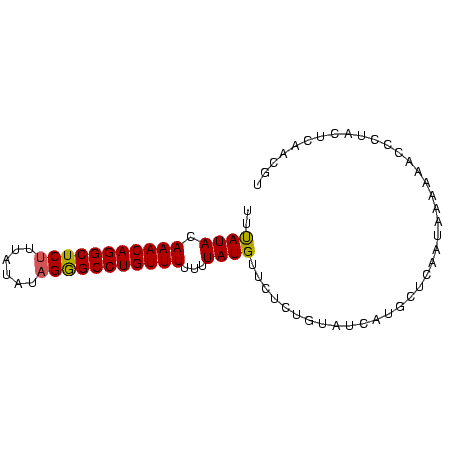
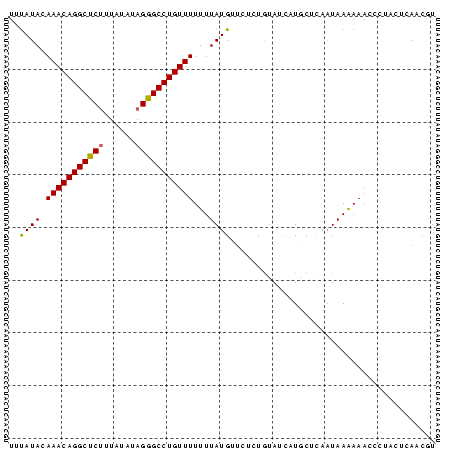
| Location | 559,018 – 559,096 |
|---|---|
| Length | 78 |
| Sequences | 3 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -12.88 |
| Energy contribution | -13.00 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 559018 78 + 4214630 ACGUUGAGUAGGGUUUUUUAUUGAGUAUGAUAUAG---ACAUAAAAAAACAGGCCCUAUAUAAAGAGCCUGUUUGUAUAAA ............((((..((((......)))).))---))(((...((((((((.((......)).)))))))).)))... ( -14.90) >Blich.0 578017 81 + 4222334 ACGUUGAGUAGGUUUUCUUAUUAAGCAUUAUACAGAGAACAUAAAAAAACAGGCUCUACAUAGAGAGCCUGUUUGUAUAAA ..(((.((((((....)))))).))).(((((((.............((((((((((......))))))))))))))))). ( -24.41) >Bclau.0 902341 81 + 4303871 ACGUUCAAGUAGGUUUUUUAGUGAGCAUGAUACAGAGAACAUAAAAAAACAGGCCCUAUUUAAGGAGCCUGUUUUUAUGAA ..((((..(((.((((......))))....)))...))))....(((((((((((((.....))).))))))))))..... ( -23.70) >consensus ACGUUGAGUAGGGUUUUUUAUUGAGCAUGAUACAGAGAACAUAAAAAAACAGGCCCUAUAUAAAGAGCCUGUUUGUAUAAA ..(((.((((((....)))))).)))....................((((((((.((......)).))))))))....... (-12.88 = -13.00 + 0.12)
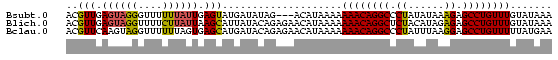
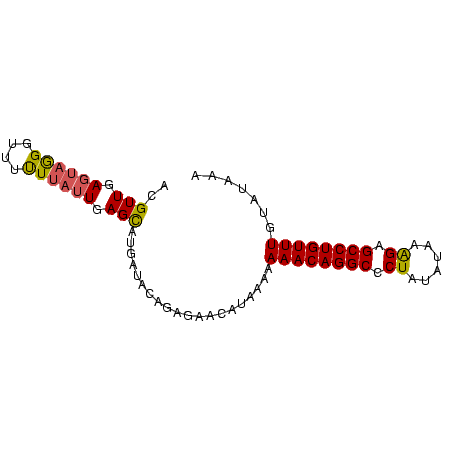
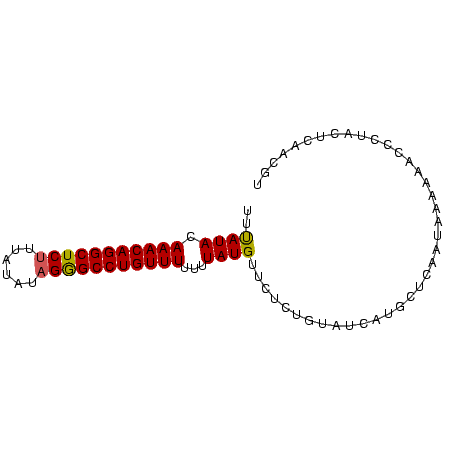
| Location | 559,018 – 559,096 |
|---|---|
| Length | 78 |
| Sequences | 3 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -18.37 |
| Consensus MFE | -15.27 |
| Energy contribution | -15.17 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.84 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 559018 78 + 4214630/0-81 UUUAUACAAACAGGCUCUUUAUAUAGGGCCUGUUUUUUUAUGU---CUAUAUCAUACUCAAUAAAAAACCCUACUCAACGU (((((..(((((((((((......)))))))))))...((((.---......))))....)))))................ ( -16.60) >Blich.0 578017 81 + 4222334/0-81 UUUAUACAAACAGGCUCUCUAUGUAGAGCCUGUUUUUUUAUGUUCUCUGUAUAAUGCUUAAUAAGAAAACCUACUCAACGU .(((((((((((((((((......)))))))))).............)))))))........................... ( -18.21) >Bclau.0 902341 81 + 4303871/0-81 UUCAUAAAAACAGGCUCCUUAAAUAGGGCCUGUUUUUUUAUGUUCUCUGUAUCAUGCUCACUAAAAAACCUACUUGAACGU .....((((((((((.(((.....)))))))))))))..((((((...(((...................)))..)))))) ( -20.31) >consensus UUUAUACAAACAGGCUCUUUAUAUAGGGCCUGUUUUUUUAUGUUCUCUGUAUCAUGCUCAAUAAAAAACCCUACUCAACGU ..((((.(((((((((((......)))))))))))...))))....................................... (-15.27 = -15.17 + -0.11)


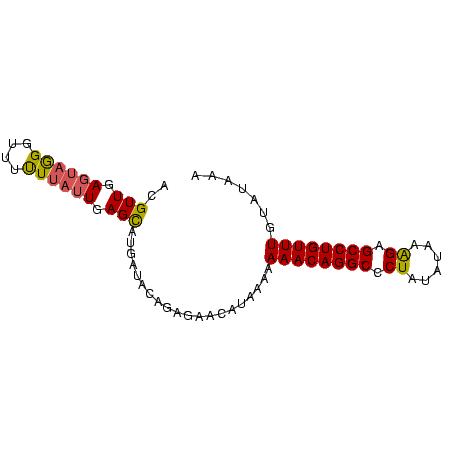
| Location | 559,018 – 559,096 |
|---|---|
| Length | 78 |
| Sequences | 3 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -12.88 |
| Energy contribution | -13.00 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 559018 78 + 4214630/0-81 ACGUUGAGUAGGGUUUUUUAUUGAGUAUGAUAUAG---ACAUAAAAAAACAGGCCCUAUAUAAAGAGCCUGUUUGUAUAAA ............((((..((((......)))).))---))(((...((((((((.((......)).)))))))).)))... ( -14.90) >Blich.0 578017 81 + 4222334/0-81 ACGUUGAGUAGGUUUUCUUAUUAAGCAUUAUACAGAGAACAUAAAAAAACAGGCUCUACAUAGAGAGCCUGUUUGUAUAAA ..(((.((((((....)))))).))).(((((((.............((((((((((......))))))))))))))))). ( -24.41) >Bclau.0 902341 81 + 4303871/0-81 ACGUUCAAGUAGGUUUUUUAGUGAGCAUGAUACAGAGAACAUAAAAAAACAGGCCCUAUUUAAGGAGCCUGUUUUUAUGAA ..((((..(((.((((......))))....)))...))))....(((((((((((((.....))).))))))))))..... ( -23.70) >consensus ACGUUGAGUAGGGUUUUUUAUUGAGCAUGAUACAGAGAACAUAAAAAAACAGGCCCUAUAUAAAGAGCCUGUUUGUAUAAA ..(((.((((((....)))))).)))....................((((((((.((......)).))))))))....... (-12.88 = -13.00 + 0.12)
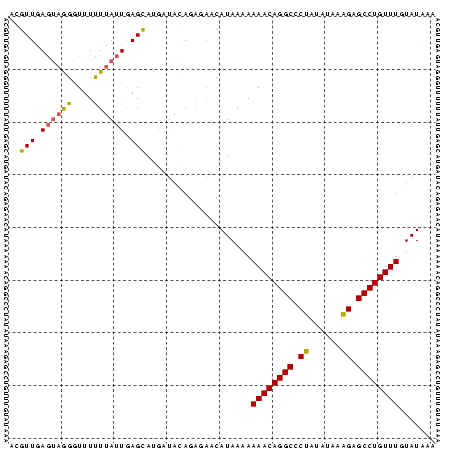
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:20:53 2006