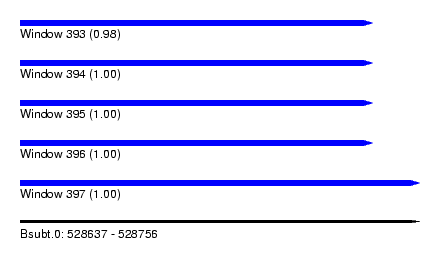
| Sequence ID | Bsubt.0 |
|---|---|
| Location | 528,637 – 528,756 |
| Length | 119 |
| Max. P | 0.999972 |

| Location | 528,637 – 528,742 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.10 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -32.33 |
| Energy contribution | -27.90 |
| Covariance contribution | -4.43 |
| Combinations/Pair | 1.48 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 528637 105 + 4214630/0-120 GAGCCAUUAGCUCAGUUGGU-AGAGCAUCUGACUUUUAAUCAGAGGGUCGAAGGUUCGAGUCCUUCAUGGCUCACCAUUUGUCUUU-------------U-UGCGGGUGUGGCGGAAUUG ..(((((..((((...((((-.((((....(((((((.....)))))))(((((.......)))))...))))))))...((....-------------.-.)))))))))))....... ( -33.80) >Blich.0 572620 106 + 4222334/0-120 GAGCCAUUAGCUCAGUUGGU-AGAGCAUCUGACUUUUAAUCAGAGGGUCGAAGGUUCGAGUCCUUCAUGGCUCACCAUUGUUUUUU-------------CACGCGGGUGUGGCGGAAUUG (((((((..((((.......-.))))....(((((((.....)))))))(((((.......))))))))))))......((((..(-------------((((....)))))..)))).. ( -34.50) >Bclau.0 891348 120 + 4303871/0-120 GGUCCGGUAGUUCAGUUGGUUAGAAUGCCUGCCUGUCACGCAGGAGGUCGCGGGUUCGAGUCCCGUCCGGACCGCCAUCUAUAUGUACAAUUAAAAUUGCCAGCGGGUGUGGCGGAAUUG (((((((...................((((.((((.....)))))))).(((((.......))))))))))))(((((((...((..((((....)))).))...)).)))))....... ( -41.30) >consensus GAGCCAUUAGCUCAGUUGGU_AGAGCAUCUGACUUUUAAUCAGAGGGUCGAAGGUUCGAGUCCUUCAUGGCUCACCAUUUAUAUUU_____________C_AGCGGGUGUGGCGGAAUUG (((((((..((((.........)))).(((((.......))))).....(((((.......)))))))))))).............................((.......))....... (-32.33 = -27.90 + -4.43)

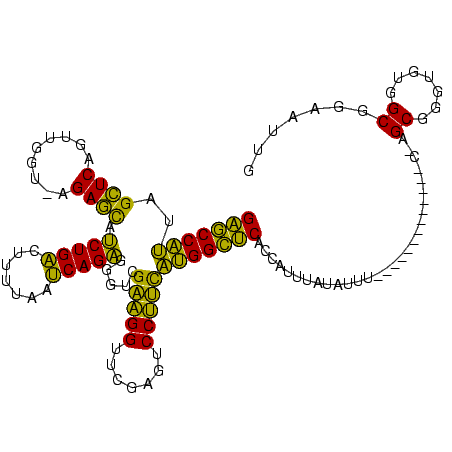
| Location | 528,637 – 528,742 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.10 |
| Mean single sequence MFE | -31.86 |
| Consensus MFE | -29.19 |
| Energy contribution | -25.98 |
| Covariance contribution | -3.21 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.92 |
| SVM decision value | 5.08 |
| SVM RNA-class probability | 0.999972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 528637 105 + 4214630/0-120 CAAUUCCGCCACACCCGCA-A-------------AAAGACAAAUGGUGAGCCAUGAAGGACUCGAACCUUCGACCCUCUGAUUAAAAGUCAGAUGCUCU-ACCAACUGAGCUAAUGGCUC .......((((......(.-.-------------...).....))))((((((((((((.......))))).....((((((.....)))))).((((.-.......))))..))))))) ( -28.20) >Blich.0 572620 106 + 4222334/0-120 CAAUUCCGCCACACCCGCGUG-------------AAAAAACAAUGGUGAGCCAUGAAGGACUCGAACCUUCGACCCUCUGAUUAAAAGUCAGAUGCUCU-ACCAACUGAGCUAAUGGCUC ...((((((.......))).)-------------))...........((((((((((((.......))))).....((((((.....)))))).((((.-.......))))..))))))) ( -30.80) >Bclau.0 891348 120 + 4303871/0-120 CAAUUCCGCCACACCCGCUGGCAAUUUUAAUUGUACAUAUAGAUGGCGGUCCGGACGGGACUCGAACCCGCGACCUCCUGCGUGACAGGCAGGCAUUCUAACCAACUGAACUACCGGACC .......((((.........(((((....))))).........))))(((((((.((((.......)))).(.(((((((.....)))).))))...................))))))) ( -36.57) >consensus CAAUUCCGCCACACCCGCA_G_____________AAAAAAAAAUGGUGAGCCAUGAAGGACUCGAACCUUCGACCCUCUGAUUAAAAGUCAGAUGCUCU_ACCAACUGAGCUAAUGGCUC .......((((................................))))((((((((((((.......))))).....((((((.....)))))).((((.........))))..))))))) (-29.19 = -25.98 + -3.21)

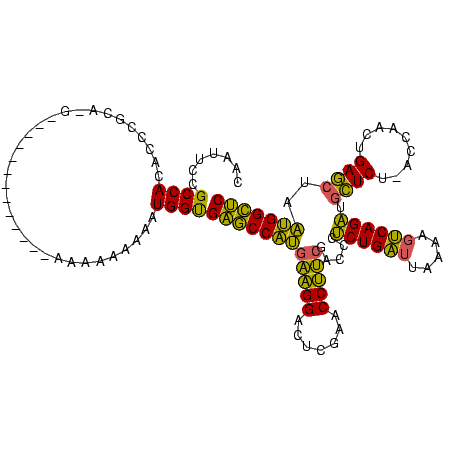
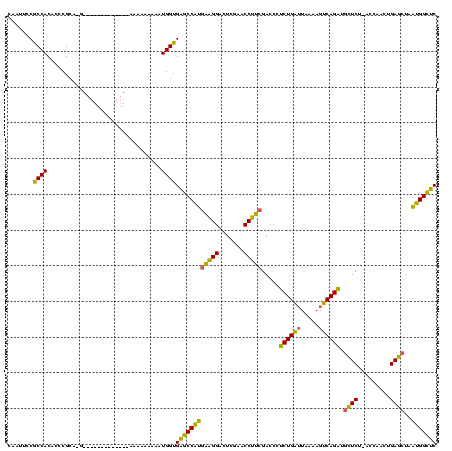
| Location | 528,637 – 528,742 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.56 |
| Mean single sequence MFE | -36.17 |
| Consensus MFE | -35.61 |
| Energy contribution | -35.17 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.84 |
| SVM RNA-class probability | 0.999654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 528637 105 + 4214630/80-200 GUCUUU-------------U-UGCGGGUGUGGCGGAAUUGGCAGACGCGCUAGACUUAGGAUCUAGUGUCUU-ACGACGUGGGGGUUCGAGUCCCUUCACCCGCACUUUAUUUACAAAGU ......-------------.-.(((((((.((.(((.(((((...((((((((((....).))))))(((..-..))))))...).)))).))))).)))))))(((((......))))) ( -35.70) >Blich.0 572620 97 + 4222334/80-200 UUUUUU-------------CACGCGGGUGUGGCGGAAUUGGCAGACGCGCUAGACUUAGGAUCUAGUGUCUUUACGGCGUGGGGGUUCGAGUCCCUUCACCCGCACUUAA---------- ......-------------...(((((((.((.(((.(((((...((((((((((((((...)))).))))....))))))...).)))).))))).)))))))......---------- ( -36.80) >Bclau.0 891348 106 + 4303871/80-200 AUAUGUACAAUUAAAAUUGCCAGCGGGUGUGGCGGAAUUGGCAGACGCGCUAGACUUAGGAUCUAGUGUCUUUAUGACGUGGGGGUUCAAGUCCCUUCACCCGCAC-------------- ......................(((((((.((.(((.(((((...((((((((((....).))))))(((.....))))))...).)))).))))).)))))))..-------------- ( -36.00) >consensus AUAUUU_____________C_AGCGGGUGUGGCGGAAUUGGCAGACGCGCUAGACUUAGGAUCUAGUGUCUUUACGACGUGGGGGUUCGAGUCCCUUCACCCGCACUU_A__________ ......................(((((((.((.(((.(((((...((((((((((....).))))))(((.....))))))...).)))).))))).)))))))................ (-35.61 = -35.17 + -0.44)

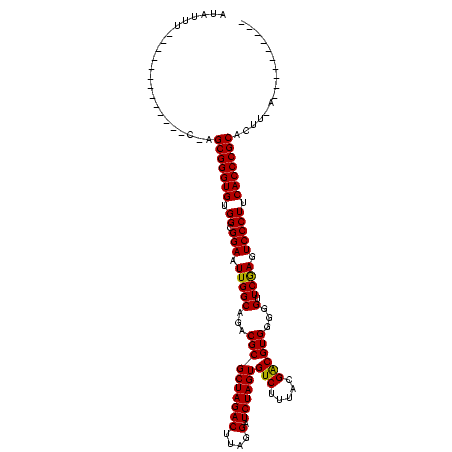
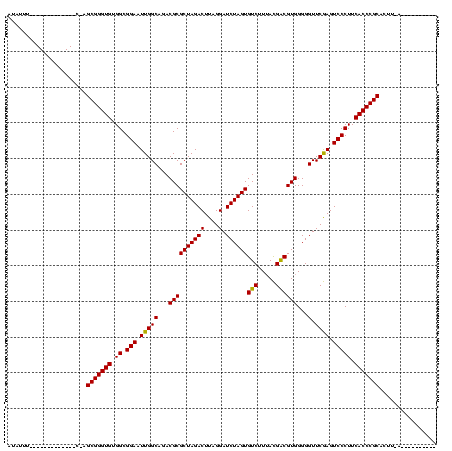
| Location | 528,637 – 528,742 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.56 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -27.25 |
| Energy contribution | -27.03 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 528637 105 + 4214630/80-200 ACUUUGUAAAUAAAGUGCGGGUGAAGGGACUCGAACCCCCACGUCGU-AAGACACUAGAUCCUAAGUCUAGCGCGUCUGCCAAUUCCGCCACACCCGCA-A-------------AAAGAC ((((((....))))))(((((((...((((.((.........(((..-..))).((((((.....)))))))).))))((.......))..))))))).-.-------------...... ( -33.00) >Blich.0 572620 97 + 4222334/80-200 ----------UUAAGUGCGGGUGAAGGGACUCGAACCCCCACGCCGUAAAGACACUAGAUCCUAAGUCUAGCGCGUCUGCCAAUUCCGCCACACCCGCGUG-------------AAAAAA ----------......(((((((..(((.........)))..((.(...((((.((((((.....))))))...)))).......).))..)))))))...-------------...... ( -27.60) >Bclau.0 891348 106 + 4303871/80-200 --------------GUGCGGGUGAAGGGACUUGAACCCCCACGUCAUAAAGACACUAGAUCCUAAGUCUAGCGCGUCUGCCAAUUCCGCCACACCCGCUGGCAAUUUUAAUUGUACAUAU --------------..(((((((..(((((.((......)).)))....((((.((((((.....))))))...)))).......))....)))))))..(((((....)))))...... ( -29.80) >consensus __________U_AAGUGCGGGUGAAGGGACUCGAACCCCCACGUCGUAAAGACACUAGAUCCUAAGUCUAGCGCGUCUGCCAAUUCCGCCACACCCGCA_G_____________AAAAAA ................(((((((..(((.........))).((..((..((((.((((((.....))))))...))))....))..))...)))))))...................... (-27.25 = -27.03 + -0.22)

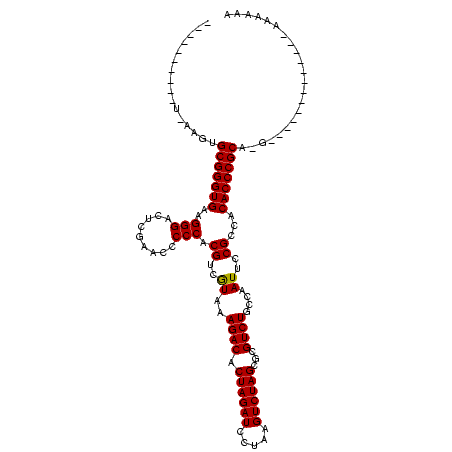
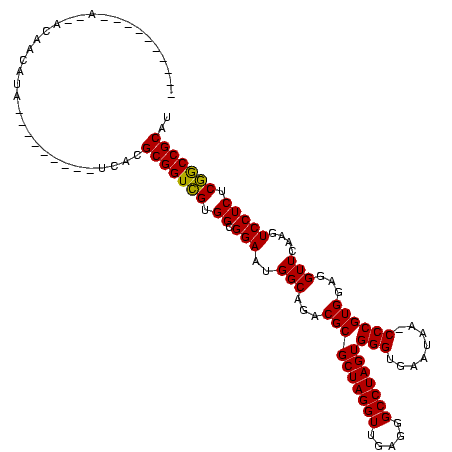
| Location | 528,637 – 528,756 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.47 |
| Mean single sequence MFE | -40.07 |
| Consensus MFE | -37.83 |
| Energy contribution | -37.17 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 528637 119 + 4214630/231-351 CUACUAAACCAGCACAACACAAUUUAAUGAUCACGCGGUCGUGGCGGAAUGGCAGACGCGCUAGGUUGAGGGCCUAGUGGGUGAAUAA-CCCGUGGAGGUUCAAGUCCUCUCGGCCGCAU ..................................(((((((.((.(((..(((...((((((((((.....)))))))((((.....)-))))))...)))....))))).))))))).. ( -40.10) >Blich.0 572620 100 + 4222334/231-351 ----------AAAACAACAUA---------UCAUGCGGUCGUGGCGGAAUGGCAGACGCGCUAGGUUGAGGGCCUAGUGGGUGAAUAA-CCCGUGGAGGUUCAAGUCCUCUCGGCCGCAU ----------...........---------..(((((((((.((.(((..(((...((((((((((.....)))))))((((.....)-))))))...)))....))))).))))))))) ( -41.10) >Bclau.0 891348 94 + 4303871/231-351 --------------CAGAAUA------------CGCGGUUGUGGCGGAAUGGCAGACGCGCUAGGUUGAGGGCCUAGUGGGAGUUAAAUCCCGUGGAGGUUCGAGUCCUCUCAACCGCAU --------------.......------------.(((((((.((.(((..(((...((((((((((.....)))))))((((......)))))))...)))....))))).))))))).. ( -39.00) >consensus __________A__ACAACAUA_________UCACGCGGUCGUGGCGGAAUGGCAGACGCGCUAGGUUGAGGGCCUAGUGGGUGAAUAA_CCCGUGGAGGUUCAAGUCCUCUCGGCCGCAU ..................................(((((((.((.(((..(((...((((((((((.....)))))))(((........))))))...)))....))))).))))))).. (-37.83 = -37.17 + -0.66)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:20:49 2006