
| Sequence ID | Banth.0 |
|---|---|
| Location | 4,530,805 – 4,530,923 |
| Length | 118 |
| Max. P | 0.999987 |

| Location | 4,530,805 – 4,530,923 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.58 |
| Mean single sequence MFE | -35.81 |
| Consensus MFE | -22.58 |
| Energy contribution | -22.70 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.22 |
| Mean z-score | -4.44 |
| Structure conservation index | 0.63 |
| SVM decision value | 4.99 |
| SVM RNA-class probability | 0.999967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4530805 118 + 5227293/0-120 ACAAAAAACCUCUCAUCCCUAGAAAGGGACGAGAGGUU--AUAGAUUCCCGCGGUACCACCCUAAAUUAAUGUACAAAAAAUAUACAUUCGCUUAGAUCCGUAACGUGGAUUAACGGCAA ......((((((((.(((((....))))).))))))))--........(((((.(((....((((.(.((((((.........)))))).).))))....))).)))))........... ( -34.10) >Bcere.0 4511843 115 + 5224283/0-120 ACAAAAAACCUCUCAUCCCUAGAAAGGGACGAGAGGUU--AUAGAUUCCCGCGGUACCACCCUAAAUUAAUGUACAA---AUAUACAUUCGCUUAGAUCCGUAACGUGGAUUAACGGCAA ......((((((((.(((((....))))).))))))))--........(((((.(((....((((.(.((((((...---...)))))).).))))....))).)))))........... ( -35.10) >Bclau.0 3001085 116 + 4303871/0-120 ACAAAAAAACUCUCUCCC---AAAAGGGACGAGAGUUUGCAUACAUUCCCGCGGUGCCACCC-ACAUUAACAGGCAUCGCCUGUUCACUUACACAUAACGGUGUGUCCCGUUAAGAGCUA ......((((((((((((---....)))).))))))))((..........((((((((....-.........))))))))...............((((((......))))))...)).. ( -38.22) >consensus ACAAAAAACCUCUCAUCCCUAGAAAGGGACGAGAGGUU__AUAGAUUCCCGCGGUACCACCCUAAAUUAAUGUACAA___AUAUACAUUCGCUUAGAUCCGUAACGUGGAUUAACGGCAA ......((((((((.(((((....))))).))))))))............((.((..............((((.......))))...........((((((.....)))))).)).)).. (-22.58 = -22.70 + 0.12)

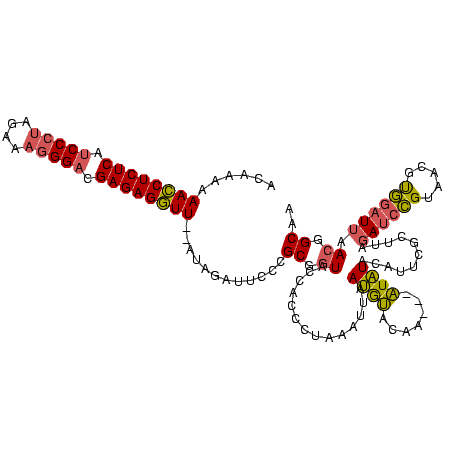
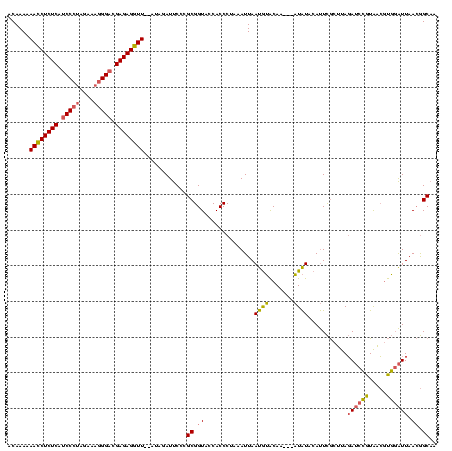
| Location | 4,530,805 – 4,530,923 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.58 |
| Mean single sequence MFE | -43.37 |
| Consensus MFE | -29.36 |
| Energy contribution | -29.27 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.27 |
| Mean z-score | -4.26 |
| Structure conservation index | 0.68 |
| SVM decision value | 5.44 |
| SVM RNA-class probability | 0.999987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4530805 118 + 5227293/0-120 UUGCCGUUAAUCCACGUUACGGAUCUAAGCGAAUGUAUAUUUUUUGUACAUUAAUUUAGGGUGGUACCGCGGGAAUCUAU--AACCUCUCGUCCCUUUCUAGGGAUGAGAGGUUUUUUGU ((.((((....((((........((((((..((((((((.....))))))))..))))))))))....)))).)).....--((((((((((((((....))))))))))))))...... ( -45.00) >Bcere.0 4511843 115 + 5224283/0-120 UUGCCGUUAAUCCACGUUACGGAUCUAAGCGAAUGUAUAU---UUGUACAUUAAUUUAGGGUGGUACCGCGGGAAUCUAU--AACCUCUCGUCCCUUUCUAGGGAUGAGAGGUUUUUUGU ((.((((....((((........((((((..(((((((..---..)))))))..))))))))))....)))).)).....--((((((((((((((....))))))))))))))...... ( -43.40) >Bclau.0 3001085 116 + 4303871/0-120 UAGCUCUUAACGGGACACACCGUUAUGUGUAAGUGAACAGGCGAUGCCUGUUAAUGU-GGGUGGCACCGCGGGAAUGUAUGCAAACUCUCGUCCCUUUU---GGGAGAGAGUUUUUUUGU .......((((((......))))))((((((....(((((((...)))))))..(((-((......)))))......))))))(((((((.((((....---)))))))))))....... ( -41.70) >consensus UUGCCGUUAAUCCACGUUACGGAUCUAAGCGAAUGUAUAU___UUGUACAUUAAUUUAGGGUGGUACCGCGGGAAUCUAU__AACCUCUCGUCCCUUUCUAGGGAUGAGAGGUUUUUUGU ...((..................((((((..((((((((.....))))))))..))))))(((....)))))..........((((((((((((((....))))))))))))))...... (-29.36 = -29.27 + -0.09)

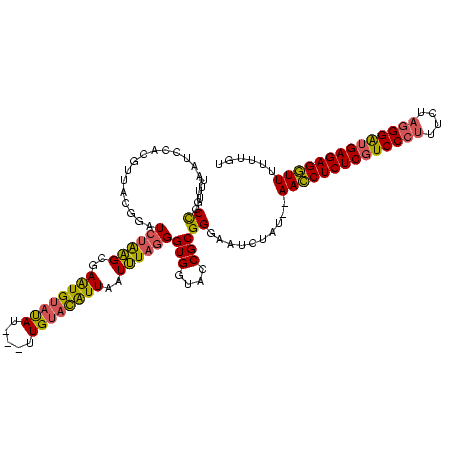
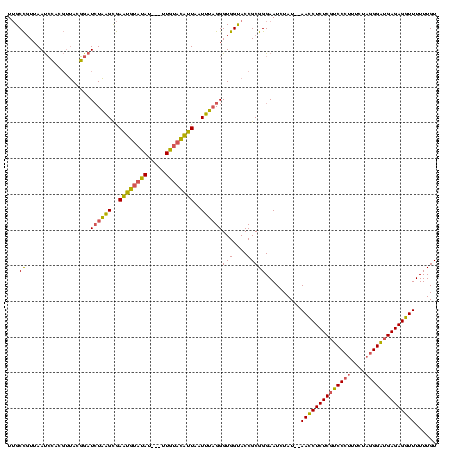
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:19:38 2006