
| Sequence ID | Banth.0 |
|---|---|
| Location | 150,577 – 2,477,513 |
| Length | 2326936 |
| Max. P | 0.999137 |

| Location | 2,477,443 – 2,477,513 |
|---|---|
| Length | 70 |
| Sequences | 3 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 92.38 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -19.53 |
| Energy contribution | -19.20 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.88 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
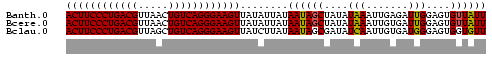
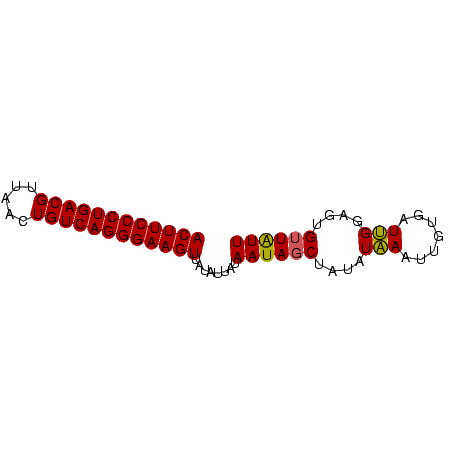
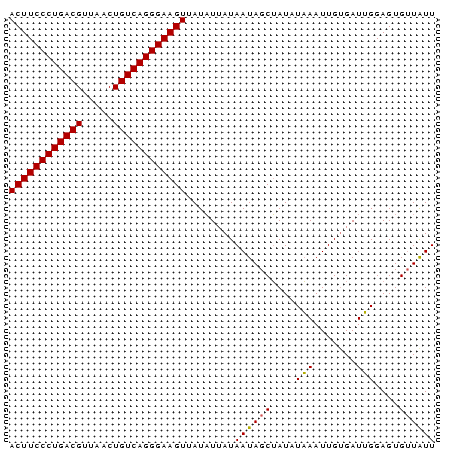
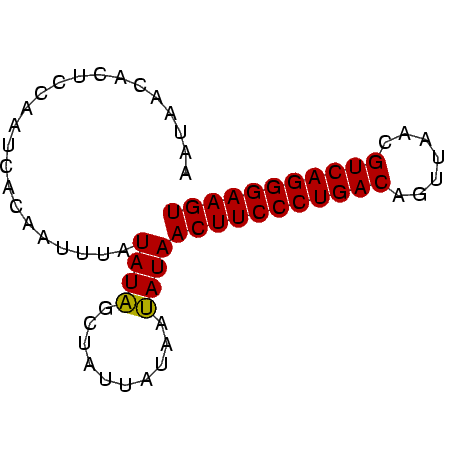
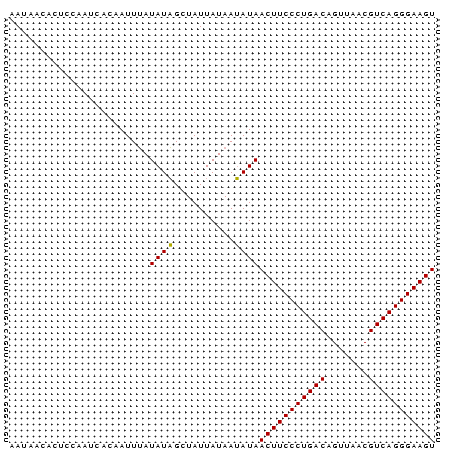
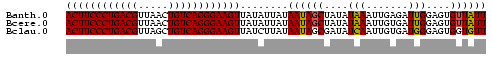
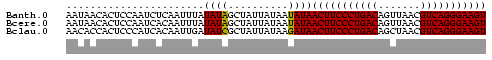
>Banth.0 2477443 70 + 5227293 ACUUCCCUGACGUUAACUGUCAGGGAAGUUAUAUUAUAAUAGCUAUAUAAAUUGAGAUUGGAGUGUUAUU ((((((((((((.....))))))))))))......((((((.(((.............)))..)))))). ( -20.02) >Bcere.0 2559648 70 + 5224283 ACUUCCCUGACGUUAACUGUCAGGGAAGUUAUAUUAUAAUAGCUAUAUAAAUUGUGAUUGGAGUGUUAUU ((((((((((((.....)))))))))))).(((((.((((.((..........)).)))).))))).... ( -20.50) >Bclau.0 426947 70 - 4303871 ACUUCCCUGACGUUAGCUGUCAGGGAAGUUAUCUUAUAAUAGCGAUAUCAAUUGUGAUGGGAGUGGUGUU ((((((((((((.....))))))))))))..((((((....(((((....))))).))))))........ ( -25.10) >consensus ACUUCCCUGACGUUAACUGUCAGGGAAGUUAUAUUAUAAUAGCUAUAUAAAUUGUGAUUGGAGUGUUAUU ((((((((((((.....))))))))))))........((((((....(((.......)))....)))))) (-19.53 = -19.20 + -0.33)
| Location | 2,477,443 – 2,477,513 |
|---|---|
| Length | 70 |
| Sequences | 3 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 92.38 |
| Mean single sequence MFE | -18.83 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.03 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -5.32 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
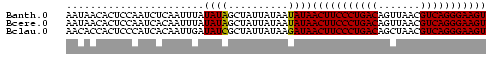
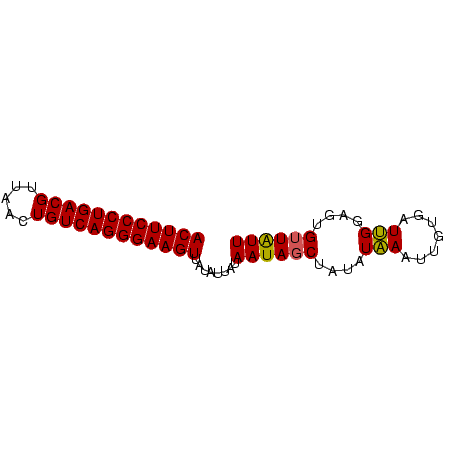
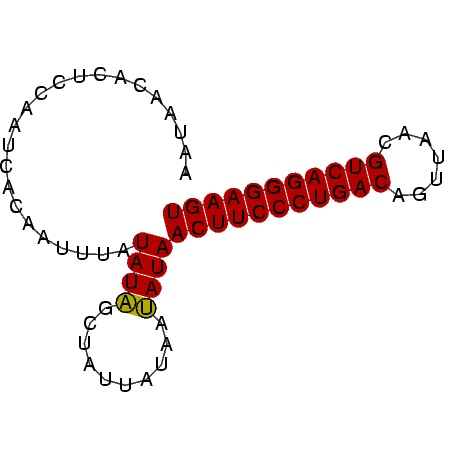
>Banth.0 2477443 70 + 5227293 AAUAACACUCCAAUCUCAAUUUAUAUAGCUAUUAUAAUAUAACUUCCCUGACAGUUAACGUCAGGGAAGU ....................(((((.......)))))....(((((((((((.......))))))))))) ( -18.50) >Bcere.0 2559648 70 + 5224283 AAUAACACUCCAAUCACAAUUUAUAUAGCUAUUAUAAUAUAACUUCCCUGACAGUUAACGUCAGGGAAGU ....................(((((.......)))))....(((((((((((.......))))))))))) ( -18.50) >Bclau.0 426947 70 - 4303871 AACACCACUCCCAUCACAAUUGAUAUCGCUAUUAUAAGAUAACUUCCCUGACAGCUAACGUCAGGGAAGU .......................((((..........))))(((((((((((.......))))))))))) ( -19.50) >consensus AAUAACACUCCAAUCACAAUUUAUAUAGCUAUUAUAAUAUAACUUCCCUGACAGUUAACGUCAGGGAAGU .......................((((..........))))(((((((((((.......))))))))))) (-18.48 = -18.03 + -0.44)
| Location | 2,477,443 – 2,477,513 |
|---|---|
| Length | 70 |
| Sequences | 3 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 92.38 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -19.53 |
| Energy contribution | -19.20 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.88 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 2477443 70 + 5227293/0-70 ACUUCCCUGACGUUAACUGUCAGGGAAGUUAUAUUAUAAUAGCUAUAUAAAUUGAGAUUGGAGUGUUAUU ((((((((((((.....))))))))))))......((((((.(((.............)))..)))))). ( -20.02) >Bcere.0 2559648 70 + 5224283/0-70 ACUUCCCUGACGUUAACUGUCAGGGAAGUUAUAUUAUAAUAGCUAUAUAAAUUGUGAUUGGAGUGUUAUU ((((((((((((.....)))))))))))).(((((.((((.((..........)).)))).))))).... ( -20.50) >Bclau.0 426947 70 - 4303871/0-70 ACUUCCCUGACGUUAGCUGUCAGGGAAGUUAUCUUAUAAUAGCGAUAUCAAUUGUGAUGGGAGUGGUGUU ((((((((((((.....))))))))))))..((((((....(((((....))))).))))))........ ( -25.10) >consensus ACUUCCCUGACGUUAACUGUCAGGGAAGUUAUAUUAUAAUAGCUAUAUAAAUUGUGAUUGGAGUGUUAUU ((((((((((((.....))))))))))))........((((((....(((.......)))....)))))) (-19.53 = -19.20 + -0.33)
| Location | 2,477,443 – 2,477,513 |
|---|---|
| Length | 70 |
| Sequences | 3 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 92.38 |
| Mean single sequence MFE | -18.83 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.03 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -5.32 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 2477443 70 + 5227293/0-70 AAUAACACUCCAAUCUCAAUUUAUAUAGCUAUUAUAAUAUAACUUCCCUGACAGUUAACGUCAGGGAAGU ....................(((((.......)))))....(((((((((((.......))))))))))) ( -18.50) >Bcere.0 2559648 70 + 5224283/0-70 AAUAACACUCCAAUCACAAUUUAUAUAGCUAUUAUAAUAUAACUUCCCUGACAGUUAACGUCAGGGAAGU ....................(((((.......)))))....(((((((((((.......))))))))))) ( -18.50) >Bclau.0 426947 70 - 4303871/0-70 AACACCACUCCCAUCACAAUUGAUAUCGCUAUUAUAAGAUAACUUCCCUGACAGCUAACGUCAGGGAAGU .......................((((..........))))(((((((((((.......))))))))))) ( -19.50) >consensus AAUAACACUCCAAUCACAAUUUAUAUAGCUAUUAUAAUAUAACUUCCCUGACAGUUAACGUCAGGGAAGU .......................((((..........))))(((((((((((.......))))))))))) (-18.48 = -18.03 + -0.44)
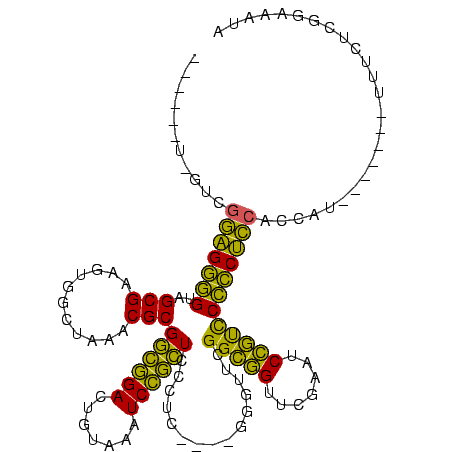
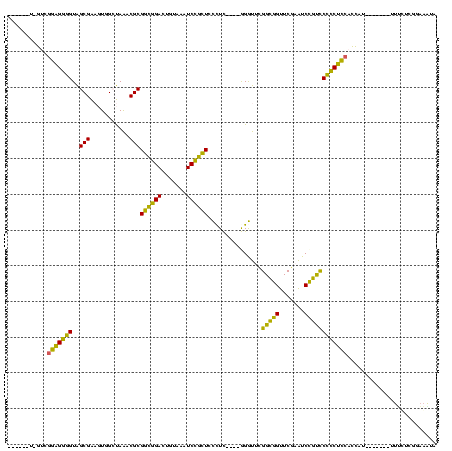
| Location | 150,577 – 150,685 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 58.46 |
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -33.24 |
| Energy contribution | -28.64 |
| Covariance contribution | -4.60 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 150577 108 + 5227293/80-200 UUAAAAU-GUCGGAGGGGUAGCGAAGUGGCUAAACGCGGCGGACUGUAAAUCCGCUCCUUC----GGGUUCGGCAGUUCGAAUCUGCCCCCCUCCACCAU-------UUUCUUUAAAAUA ..(((((-(..(((((((((((......))))...((((((((((((.((((((......)----)))))..))))))))...)))).)))))))..)))-------))).......... ( -43.10) >Bcere.0 150583 108 + 5224283/80-200 UUUAAAU-GUCGGAGGGGUAGCGAAGUGGCUAAACGCGGCGGACUGUAAAUCCGCUCCUUC----GGGUUCGGCAGUUCGAAUCUGCCCCCCUCCACCAU-------UUUCUUUAAAAUA ...((((-(..(((((((((((......))))...((((((((((((.((((((......)----)))))..))))))))...)))).)))))))..)))-------))........... ( -42.20) >Bhalo.0 282564 113 + 4202352/80-200 CACGCUUCGAUGGAGGGGUAGCGAAGUGGCUAAACGCGGCGGACUGUAAAUCCGCUCCCUCA---GGGUUCGGCGGUUCGAAUCCGUCCCCCUCCACCAUGGA----UUGAUGGGCUAUA ...((.....((((((((((((......))))...((((((((((((.(((((.........---)))))..))))))))...)))).))))))))((((...----...)))))).... ( -43.10) >Bsubt.0 1042010 104 - 4214630/80-200 --------GUUGCGGAUGUGGCGGAAUUGGCAGACGCGCUAGAAUCAGGCUCUAGUGUCUUU-ACAGACGUGGGGGUUCAAGUCCCUUCAUCCGCACCAU-------UUCUGCGGAAGUA --------(.((((((((.((.(((.(((((...(((((((((.......))))))((((..-..)))))))...).)))).))))).)))))))).)..-------...(((....))) ( -37.70) >Blich.0 1105229 109 - 4222334/80-200 --------G---CGGAUGUGGCGGAAUUGGCAGACGCGCUAGAAUCAGGCUCUAGUGUCUUUUAAAGACGUGGGGGUUCAAGUCCCUUCAUCCGCAUUGCCAAUCUAUUUUGCGGAAGUA --------(---((((((.((.(((.(((((...(((((((((.......))))))((((.....)))))))...).)))).))))).)))))))...............(((....))) ( -37.00) >consensus ______U_GUCGGAGGGGUAGCGAAGUGGCUAAACGCGGCGGACUGUAAAUCCGCUCCCUC____GGGUUCGGCGGUUCGAAUCCGUCCCCCUCCACCAU_______UUUCUCGGAAAUA ...........(((((((..(((...........)))((((((.......))))))...............(((((.......))))))))))))......................... (-33.24 = -28.64 + -4.60)

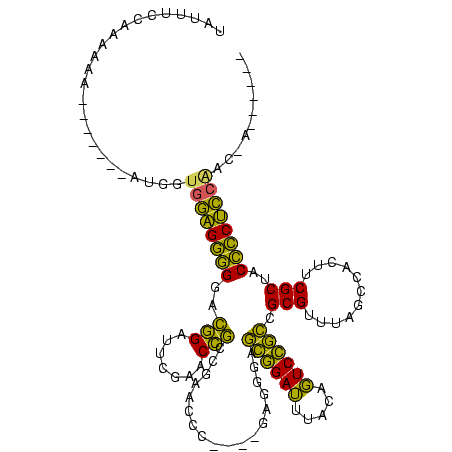
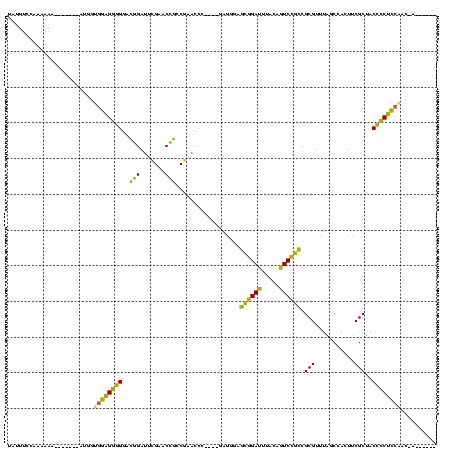
| Location | 150,577 – 150,685 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 58.46 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -28.98 |
| Energy contribution | -24.42 |
| Covariance contribution | -4.56 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 150577 108 + 5227293/80-200 UAUUUUAAAGAAA-------AUGGUGGAGGGGGGCAGAUUCGAACUGCCGAACCC----GAAGGAGCGGAUUUACAGUCCGCCGCGUUUAGCCACUUCGCUACCCCUCCGAC-AUUUUAA ..........(((-------(((.(((((((((((((.......))))).....(----(((((.((((((.....)))))))((.....))..)))))...)))))))).)-))))).. ( -45.40) >Bcere.0 150583 108 + 5224283/80-200 UAUUUUAAAGAAA-------AUGGUGGAGGGGGGCAGAUUCGAACUGCCGAACCC----GAAGGAGCGGAUUUACAGUCCGCCGCGUUUAGCCACUUCGCUACCCCUCCGAC-AUUUAAA ...........((-------(((.(((((((((((((.......))))).....(----(((((.((((((.....)))))))((.....))..)))))...)))))))).)-))))... ( -44.40) >Bhalo.0 282564 113 + 4202352/80-200 UAUAGCCCAUCAA----UCCAUGGUGGAGGGGGACGGAUUCGAACCGCCGAACCC---UGAGGGAGCGGAUUUACAGUCCGCCGCGUUUAGCCACUUCGCUACCCCUCCAUCGAAGCGUG .............----....((((((((((((((((.((((......)))).))---...(.(.((((((.....))))))).))))((((......)))))))))))))))....... ( -41.80) >Bsubt.0 1042010 104 - 4214630/80-200 UACUUCCGCAGAA-------AUGGUGCGGAUGAAGGGACUUGAACCCCCACGUCUGU-AAAGACACUAGAGCCUGAUUCUAGCGCGUCUGCCAAUUCCGCCACAUCCGCAAC-------- .......((.(((-------.(((((((((((..(((........)))..)))))))-..((((.((((((.....))))))...)))))))).))).))............-------- ( -29.70) >Blich.0 1105229 109 - 4222334/80-200 UACUUCCGCAAAAUAGAUUGGCAAUGCGGAUGAAGGGACUUGAACCCCCACGUCUUUAAAAGACACUAGAGCCUGAUUCUAGCGCGUCUGCCAAUUCCGCCACAUCCG---C-------- ...............(((((((((((((((((..(((........)))..))))...........((((((.....))))))))))).))))))))............---.-------- ( -28.20) >consensus UAUUUCCAAAAAA_______AUGGUGGAGGGGGACGGAUUCGAACCGCCGAACCC____GAGGGAGCGGAUUUACAGUCCGCCGCGUUUAGCCACUUCGCUACCCCUCCAAC_A______ ........................((((((((..(((.......)))..................((((((.....)))))).(((...........)))..)))))))).......... (-28.98 = -24.42 + -4.56)

| Location | 150,577 – 150,683 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 54.40 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -13.10 |
| Energy contribution | -9.18 |
| Covariance contribution | -3.92 |
| Combinations/Pair | 1.68 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 150577 106 + 5227293/120-240 GGACUGUAAAUCCGCUCCUUC----GGGUUCGGCAGUUCGAAUCUGCCCCCCUCCACCAU-------UUUCUUUAAAAUAACC---UGAGAGGGUUUAUUUUUUUGAAAAGAUAACCAUA (((.....((((((......)----))))).(((((.......)))))....)))....(-------((((...(((((((((---(....))).)))))))...))))).......... ( -26.90) >Bcere.0 150583 106 + 5224283/120-240 GGACUGUAAAUCCGCUCCUUC----GGGUUCGGCAGUUCGAAUCUGCCCCCCUCCACCAU-------UUUCUUUAAAAUAACC---UGAGAGGGUUUAUUUUUUUGAAAAGAUAACCAUA (((.....((((((......)----))))).(((((.......)))))....)))....(-------((((...(((((((((---(....))).)))))))...))))).......... ( -26.90) >Bhalo.0 282564 112 + 4202352/120-240 GGACUGUAAAUCCGCUCCCUCA---GGGUUCGGCGGUUCGAAUCCGUCCCCCUCCACCAUGGA----UUGAUGGGCUAUAGCCAAGCGGUAAGGCAACGGACUUUGACUCCGUGAU-GCG ....(((....(((((.(.(((---(((...(((((.......))))).)))(((.....)))----.))).)(((....))).)))))....)))(((((.......)))))...-... ( -33.50) >Bsubt.0 1042010 110 - 4214630/120-240 AGAAUCAGGCUCUAGUGUCUUU-ACAGACGUGGGGGUUCAAGUCCCUUCAUCCGCACCAU-------UUCUGCGGAAGUAGUUCAGUGGUAGAAC--ACCACCUUGCCAAGGUGGGGGUC .......(((((..((((((..-..))))(((((((.......))).....))))(((((-------(.((((....))))...))))))...))--.((((((.....))))))))))) ( -36.40) >Blich.0 1105229 118 - 4222334/120-240 AGAAUCAGGCUCUAGUGUCUUUUAAAGACGUGGGGGUUCAAGUCCCUUCAUCCGCAUUGCCAAUCUAUUUUGCGGAAGUAGUUCAGUGGUAGAAC--ACCACCUUGCCAAGGUGGGGGUC .......((((((..(((((.....)))))..))))))...(.(((....((((((..............))))))....((((.......))))--.((((((.....))))))))).) ( -34.94) >consensus GGACUGUAAAUCCGCUCCCUC____GGGUUCGGCGGUUCGAAUCCGUCCCCCUCCACCAU_______UUUCUCGGAAAUAGCC_AGUGGUAGGGC__ACCACUUUGACAAGGUGACCGUA (((.......))).............((...(((((.......)))))..))............................(((.........)))...((((((.....))))))..... (-13.10 = -9.18 + -3.92)

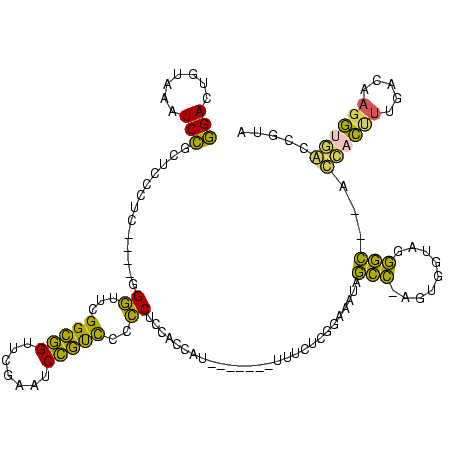
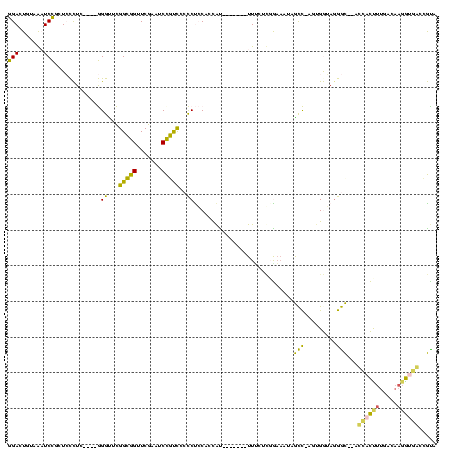
| Location | 150,577 – 150,685 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.88 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -29.24 |
| Energy contribution | -26.12 |
| Covariance contribution | -3.12 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 150577 108 + 5227293/360-480 UAGCCCAGCC-AUUUACGAGCCAUUAGCUCAGUUGG-UAGAGCAUCUGACUUUUAAUCAGAGGGUCGAAGGUUCGAGUCCUUCAUGGCUCACCAU----UU--UCG----CGGAAGUAGU ....((.((.-......((((.....))))...(((-(.((((....(((((((.....)))))))(((((.......)))))...)))))))).----..--..)----)))....... ( -31.70) >Bcere.0 150583 108 + 5224283/360-480 UAGCCCAGCC-AUUUACGAGCCAUUAGCUCAGUUGG-UAGAGCAUCUGACUUUUAAUCAGAGGGUCGAAGGUUCGAGUCCUUCAUGGCUCACCAU----UU--UCG----CGGAAGUAGU ....((.((.-......((((.....))))...(((-(.((((....(((((((.....)))))))(((((.......)))))...)))))))).----..--..)----)))....... ( -31.70) >Bhalo.0 282564 111 + 4202352/360-480 UGAUCUUUUUUGUUUUAGAGCCAUUAGCUCAGUUGG-UAGAGCAUCUGACUUUUAAUCAGAGGGUCGAAGGUUCGAGUCCUUCAUGGCUCACCAU----GUCAUUG----CGGAAGUAGU .(((.............((((.....))))...(((-(.((((....(((((((.....)))))))(((((.......)))))...)))))))).----)))((((----(....))))) ( -34.00) >Bsubt.0 1042010 120 - 4214630/360-480 CCUCGGCAUUAAGUUUUGCGCCCGUAGCUCAAUUGGAUAGAGCGUUUGACUACGGAUCAAAAGGUUAGGGGUUCGACUCCUCUCGGGCGCGCCAUGAUCUAUAUGAAAUCGGGAAGUAGC .(((((...........(((((((..((((.........)))).((((((....).))))).....(((((.....)))))..)))))))..((((.....))))...)))))....... ( -33.90) >Blich.0 1105229 96 - 4222334/360-480 -----------------GCGCCCGUAGCUCAAUUGGAUAGAGCGUUUGACUACGGAUCAAAAGGUUAGGGGUUCGACUCCUCUCGGGCGCGCCAU----UAUAUAA---CGGGAAGUAGC -----------------(((((((..((((.........)))).((((((....).))))).....(((((.....)))))..))))))).....----.......---........... ( -29.50) >consensus UAGCCCAGCC_AUUUACGAGCCAUUAGCUCAGUUGG_UAGAGCAUCUGACUUUUAAUCAGAGGGUCGAAGGUUCGAGUCCUUCAUGGCUCACCAU____UU_AUCG____CGGAAGUAGU .................(((((((..((((.........)))).(((((.......))))).....(((((.......)))))))))))).............................. (-29.24 = -26.12 + -3.12)

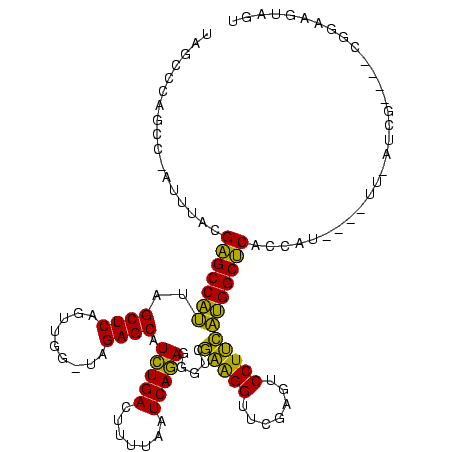
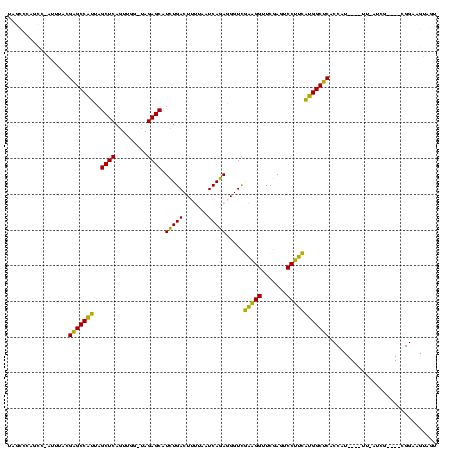
| Location | 150,577 – 150,685 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.88 |
| Mean single sequence MFE | -30.56 |
| Consensus MFE | -28.74 |
| Energy contribution | -26.26 |
| Covariance contribution | -2.48 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 150577 108 + 5227293/360-480 ACUACUUCCG----CGA--AA----AUGGUGAGCCAUGAAGGACUCGAACCUUCGACCCUCUGAUUAAAAGUCAGAUGCUCUA-CCAACUGAGCUAAUGGCUCGUAAAU-GGCUGGGCUA ......((((----(..--..----.((((((((..((((((.......))))))....((((((.....)))))).)))).)-)))...((((.....))))......-.)).)))... ( -32.00) >Bcere.0 150583 108 + 5224283/360-480 ACUACUUCCG----CGA--AA----AUGGUGAGCCAUGAAGGACUCGAACCUUCGACCCUCUGAUUAAAAGUCAGAUGCUCUA-CCAACUGAGCUAAUGGCUCGUAAAU-GGCUGGGCUA ......((((----(..--..----.((((((((..((((((.......))))))....((((((.....)))))).)))).)-)))...((((.....))))......-.)).)))... ( -32.00) >Bhalo.0 282564 111 + 4202352/360-480 ACUACUUCCG----CAAUGAC----AUGGUGAGCCAUGAAGGACUCGAACCUUCGACCCUCUGAUUAAAAGUCAGAUGCUCUA-CCAACUGAGCUAAUGGCUCUAAAACAAAAAAGAUCA ((((..((..----....)).----.))))((((((((((((.......))))).....((((((.....)))))).((((..-......))))..)))))))................. ( -28.90) >Bsubt.0 1042010 120 - 4214630/360-480 GCUACUUCCCGAUUUCAUAUAGAUCAUGGCGCGCCCGAGAGGAGUCGAACCCCUAACCUUUUGAUCCGUAGUCAAACGCUCUAUCCAAUUGAGCUACGGGCGCAAAACUUAAUGCCGAGG ....((((..(((((.....)))))..((((((((((...(((.(((((..........))))))))..........((((.........))))..)))))))..........))))))) ( -32.20) >Blich.0 1105229 96 - 4222334/360-480 GCUACUUCCCG---UUAUAUA----AUGGCGCGCCCGAGAGGAGUCGAACCCCUAACCUUUUGAUCCGUAGUCAAACGCUCUAUCCAAUUGAGCUACGGGCGC----------------- ........(((---((....)----)))).(((((((...(((.(((((..........))))))))..........((((.........))))..)))))))----------------- ( -27.70) >consensus ACUACUUCCG____CGAU_AA____AUGGUGAGCCAUGAAGGACUCGAACCUUCGACCCUCUGAUUAAAAGUCAGAUGCUCUA_CCAACUGAGCUAAUGGCUCGAAAAU_AACUGGGCUA ..............................((((((((((((.......))))).....((((((.....)))))).((((.........))))..)))))))................. (-28.74 = -26.26 + -2.48)

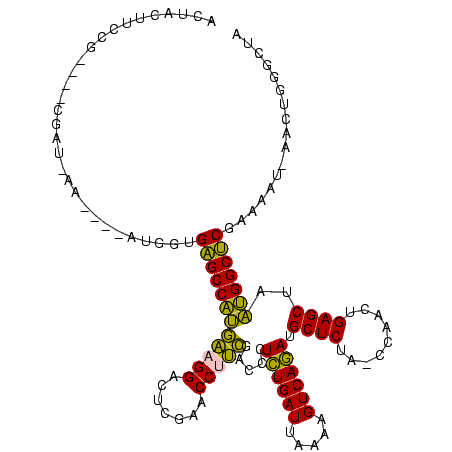
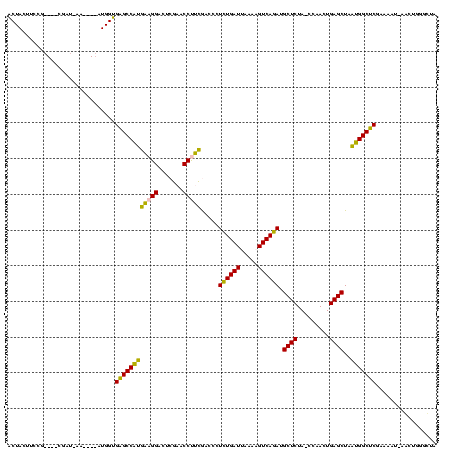
| Location | 150,577 – 150,685 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.63 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -28.30 |
| Energy contribution | -24.22 |
| Covariance contribution | -4.08 |
| Combinations/Pair | 1.42 |
| Mean z-score | -0.56 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 150577 108 + 5227293/400-520 AGCAUCUGACUUUUAAUCAGAGGGUCGAAGGUUCGAGUCCUUCAUGGCUCACCAU----UU--UCG----CGGAAGUAGUUCAG--UGGUAGAAUACAACCUUGCCAAGGUUGGGGUCGC .((....(((((((.....)))))))(((((.......)))))..((((((((((----(.--(..----(....)..)...))--))))......((((((.....))))))))))))) ( -33.90) >Bcere.0 150583 108 + 5224283/400-520 AGCAUCUGACUUUUAAUCAGAGGGUCGAAGGUUCGAGUCCUUCAUGGCUCACCAU----UU--UCG----CGGAAGUAGUUCAG--UGGUAGAAUACAACCUUGCCAAGGUUGGGGUCGC .((....(((((((.....)))))))(((((.......)))))..((((((((((----(.--(..----(....)..)...))--))))......((((((.....))))))))))))) ( -33.90) >Bhalo.0 282564 110 + 4202352/400-520 AGCAUCUGACUUUUAAUCAGAGGGUCGAAGGUUCGAGUCCUUCAUGGCUCACCAU----GUCAUUG----CGGAAGUAGUUCAG--UGGUAGAACACCACCUUGCCAAGGUGGGGGUCGC .((....(((((((.....)))))))(((((.......)))))..((((((((((----...((((----(....)))))...)--))))......((((((.....))))))))))))) ( -40.80) >Bsubt.0 1042010 120 - 4214630/400-520 AGCGUUUGACUACGGAUCAAAAGGUUAGGGGUUCGACUCCUCUCGGGCGCGCCAUGAUCUAUAUGAAAUCGGGAAGUAGCUCAGCUUGGUAGAGCACAUGGUUUGGGACCAUGGGGUCGC .((((((((.....((((....))))(((((.....))))).))))))))((((((..............(((......))).((((....)))).))))))....((((....)))).. ( -35.10) >Blich.0 1105229 113 - 4222334/400-520 AGCGUUUGACUACGGAUCAAAAGGUUAGGGGUUCGACUCCUCUCGGGCGCGCCAU----UAUAUAA---CGGGAAGUAGCUCAGCUUGGUAGAGCACAUGGUUUGGGACCAUGGGGUCGC .((((((((.....((((....))))(((((.....))))).)))))))).....----.......---......(..((((.((((....)))).(((((((...)))))))))))..) ( -32.80) >consensus AGCAUCUGACUUUUAAUCAGAGGGUCGAAGGUUCGAGUCCUUCAUGGCUCACCAU____UU_AUCG____CGGAAGUAGUUCAG__UGGUAGAACACAACCUUGCCAAGGUUGGGGUCGC .((.(((((.......)))))((((((((((.......)))))..)))))(((.........................((((.........)))).(((((((...))))))).))).)) (-28.30 = -24.22 + -4.08)

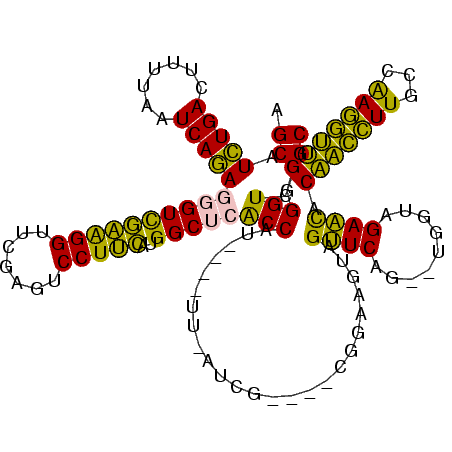
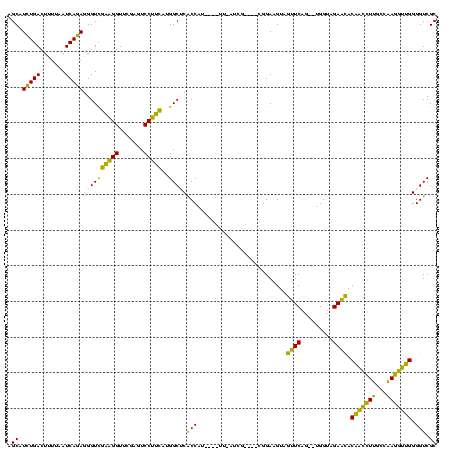
| Location | 150,577 – 150,685 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.63 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -22.72 |
| Energy contribution | -20.00 |
| Covariance contribution | -2.72 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.12 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 150577 108 + 5227293/400-520 GCGACCCCAACCUUGGCAAGGUUGUAUUCUACCA--CUGAACUACUUCCG----CGA--AA----AUGGUGAGCCAUGAAGGACUCGAACCUUCGACCCUCUGAUUAAAAGUCAGAUGCU (((....((((((.....))))))..(((.....--..))).......))----)..--..----((((....))))(((((.......))))).....((((((.....)))))).... ( -25.00) >Bcere.0 150583 108 + 5224283/400-520 GCGACCCCAACCUUGGCAAGGUUGUAUUCUACCA--CUGAACUACUUCCG----CGA--AA----AUGGUGAGCCAUGAAGGACUCGAACCUUCGACCCUCUGAUUAAAAGUCAGAUGCU (((....((((((.....))))))..(((.....--..))).......))----)..--..----((((....))))(((((.......))))).....((((((.....)))))).... ( -25.00) >Bhalo.0 282564 110 + 4202352/400-520 GCGACCCCCACCUUGGCAAGGUGGUGUUCUACCA--CUGAACUACUUCCG----CAAUGAC----AUGGUGAGCCAUGAAGGACUCGAACCUUCGACCCUCUGAUUAAAAGUCAGAUGCU (((....((((((.....)))))).((((.....--..))))......))----).....(----((((....)))))..((..((((....)))).))((((((.....)))))).... ( -31.70) >Bsubt.0 1042010 120 - 4214630/400-520 GCGACCCCAUGGUCCCAAACCAUGUGCUCUACCAAGCUGAGCUACUUCCCGAUUUCAUAUAGAUCAUGGCGCGCCCGAGAGGAGUCGAACCCCUAACCUUUUGAUCCGUAGUCAAACGCU (((....((((((.....)))))).((((.........))))........(((((.....))))).(((((((..(((((((..............)))))))...))).))))..))). ( -27.14) >Blich.0 1105229 113 - 4222334/400-520 GCGACCCCAUGGUCCCAAACCAUGUGCUCUACCAAGCUGAGCUACUUCCCG---UUAUAUA----AUGGCGCGCCCGAGAGGAGUCGAACCCCUAACCUUUUGAUCCGUAGUCAAACGCU (((....((((((.....)))))).((((.........)))).........---.......----.(((((((..(((((((..............)))))))...))).))))..))). ( -25.74) >consensus GCGACCCCAACCUUGGCAAGGUUGUGUUCUACCA__CUGAACUACUUCCG____CGAU_AA____AUGGUGAGCCAUGAAGGACUCGAACCUUCGACCCUCUGAUUAAAAGUCAGAUGCU ((((((.((((((.....)))))).((((.........)))).........................))).......(((((.......))))).....((((((.....))))))))). (-22.72 = -20.00 + -2.72)

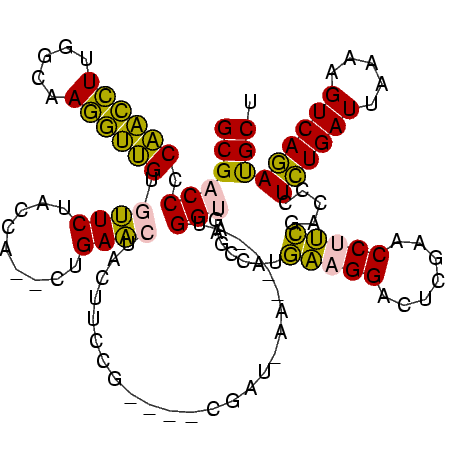
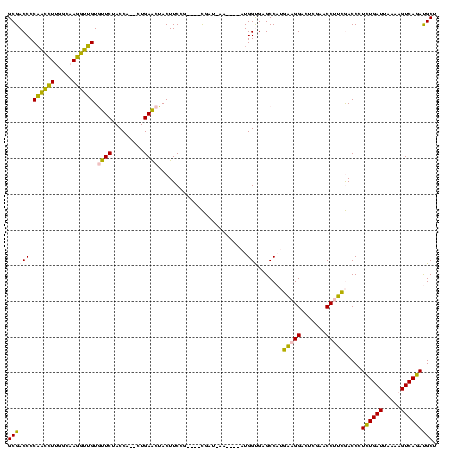
| Location | 150,577 – 150,685 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.73 |
| Mean single sequence MFE | -40.24 |
| Consensus MFE | -31.69 |
| Energy contribution | -28.01 |
| Covariance contribution | -3.68 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 150577 108 + 5227293/440-560 UUCAUGGCUCACCAU----UU--UCG----CGGAAGUAGUUCAG--UGGUAGAAUACAACCUUGCCAAGGUUGGGGUCGCGGGUUCGAAUCCCGUCUUCCGCUUAAUUACUAAGGGGCCU .....(((((.((..----..--...----.)).(((((((.((--(((.(((...((((((.....))))))(((...((....))...))).))).))))).)))))))...))))). ( -38.50) >Bcere.0 150583 108 + 5224283/440-560 UUCAUGGCUCACCAU----UU--UCG----CGGAAGUAGUUCAG--UGGUAGAAUACAACCUUGCCAAGGUUGGGGUCGCGGGUUCGAAUCCCGUCUUCCGCUUAAUUACUAAGGGGCCU .....(((((.((..----..--...----.)).(((((((.((--(((.(((...((((((.....))))))(((...((....))...))).))).))))).)))))))...))))). ( -38.50) >Bhalo.0 282564 107 + 4202352/440-560 UUCAUGGCUCACCAU----GUCAUUG----CGGAAGUAGUUCAG--UGGUAGAACACCACCUUGCCAAGGUGGGGGUCGCGGGUUCGAAUCCCGUCUUCCGCUC---CAAAAAGGGGCCU .....((((((((((----...((((----(....)))))...)--))))......((((((.....)))))))))))(((((.......))))).....((((---(.....))))).. ( -42.50) >Bsubt.0 1042010 118 - 4214630/440-560 UCUCGGGCGCGCCAUGAUCUAUAUGAAAUCGGGAAGUAGCUCAGCUUGGUAGAGCACAUGGUUUGGGACCAUGGGGUCGCAGGUUCGAAUCCUGUCUUCCCGACCAUUCUU--GGGGCCU ....((((.(.(((.((((.....))..((((((((..((((.((((....)))).(((((((...))))))))))).(((((.......)))))))))))))....)).)--))))))) ( -42.80) >Blich.0 1105229 112 - 4222334/440-560 UCUCGGGCGCGCCAU----UAUAUAA---CGGGAAGUAGCUCAGCUUGGUAGAGCACAUGGUUUGGGACCAUGGGGUCGCAGGUUCGAAUCCUGUCUUCCCGACCACUAUUC-GGGGCCU ....((((.......----.......---(((((((..((((.((((....)))).(((((((...))))))))))).(((((.......)))))))))))).((.......-.)))))) ( -38.90) >consensus UUCAUGGCUCACCAU____UU_AUCG____CGGAAGUAGUUCAG__UGGUAGAACACAACCUUGCCAAGGUUGGGGUCGCGGGUUCGAAUCCCGUCUUCCGCUCAAUUACUAAGGGGCCU .....(((.(.((.................((((((..((((.........)))).(((((((...))))))).....(((((.......)))))))))))............)))))). (-31.69 = -28.01 + -3.68)

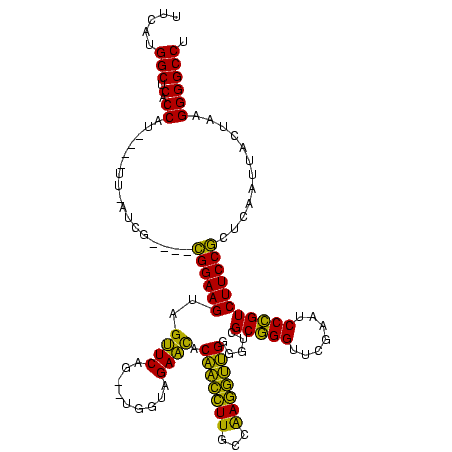
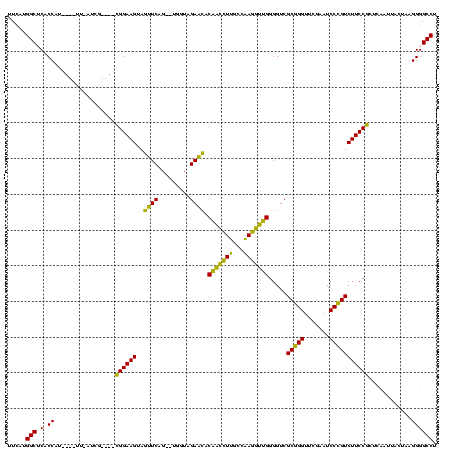
| Location | 150,577 – 150,685 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.73 |
| Mean single sequence MFE | -34.23 |
| Consensus MFE | -28.59 |
| Energy contribution | -25.79 |
| Covariance contribution | -2.80 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 150577 108 + 5227293/440-560 AGGCCCCUUAGUAAUUAAGCGGAAGACGGGAUUCGAACCCGCGACCCCAACCUUGGCAAGGUUGUAUUCUACCA--CUGAACUACUUCCG----CGA--AA----AUGGUGAGCCAUGAA .((((((...........(((((((..(((.......)))(((((((((....)))...)))))).........--........))))))----)..--..----..)).).)))..... ( -30.53) >Bcere.0 150583 108 + 5224283/440-560 AGGCCCCUUAGUAAUUAAGCGGAAGACGGGAUUCGAACCCGCGACCCCAACCUUGGCAAGGUUGUAUUCUACCA--CUGAACUACUUCCG----CGA--AA----AUGGUGAGCCAUGAA .((((((...........(((((((..(((.......)))(((((((((....)))...)))))).........--........))))))----)..--..----..)).).)))..... ( -30.53) >Bhalo.0 282564 107 + 4202352/440-560 AGGCCCCUUUUUG---GAGCGGAAGACGGGAUUCGAACCCGCGACCCCCACCUUGGCAAGGUGGUGUUCUACCA--CUGAACUACUUCCG----CAAUGAC----AUGGUGAGCCAUGAA .....((.....)---).(((((((..(((..(((......)))..)))....(((...(((((((...)))))--))...)))))))))----).....(----((((....))))).. ( -37.50) >Bsubt.0 1042010 118 - 4214630/440-560 AGGCCCC--AAGAAUGGUCGGGAAGACAGGAUUCGAACCUGCGACCCCAUGGUCCCAAACCAUGUGCUCUACCAAGCUGAGCUACUUCCCGAUUUCAUAUAGAUCAUGGCGCGCCCGAGA .((((((--(.((((((((((((((.((((.......))))......((((((.....)))))).((((.........))))..)))))))))..))).....)).))).).)))..... ( -37.70) >Blich.0 1105229 112 - 4222334/440-560 AGGCCCC-GAAUAGUGGUCGGGAAGACAGGAUUCGAACCUGCGACCCCAUGGUCCCAAACCAUGUGCUCUACCAAGCUGAGCUACUUCCCG---UUAUAUA----AUGGCGCGCCCGAGA .((((((-(....((((.(((((((.((((.......))))......((((((.....)))))).((((.........))))..)))))))---))))...----.))).).)))..... ( -34.90) >consensus AGGCCCCUUAAUAAUGGAGCGGAAGACGGGAUUCGAACCCGCGACCCCAACCUUGGCAAGGUUGUGUUCUACCA__CUGAACUACUUCCG____CGAU_AA____AUGGUGAGCCAUGAA .((((((............((((((.((((.......))))......((((((.....)))))).((((.........))))..)))))).................)).).)))..... (-28.59 = -25.79 + -2.80)

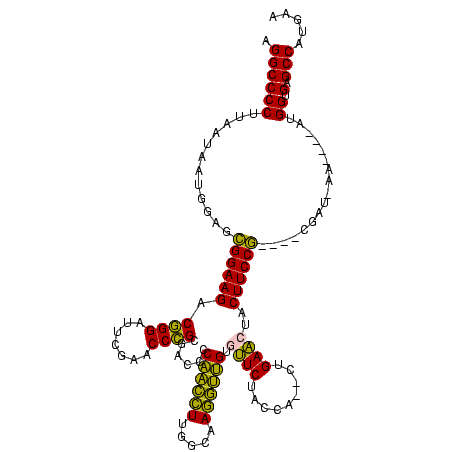
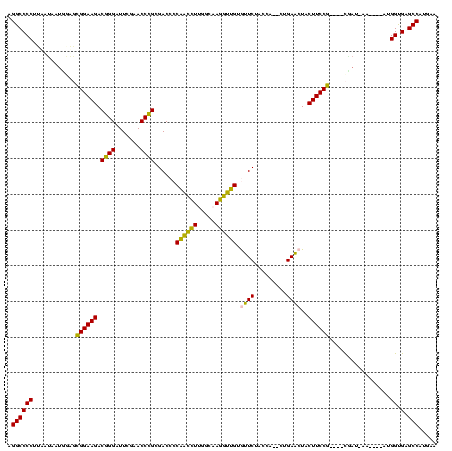
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:19:35 2006