| Sequence ID | Banth.0 |
|---|---|
| Location | 150,318 – 150,410 |
| Length | 92 |
| Max. P | 0.974138 |

| Location | 150,318 – 150,409 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.58 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -33.75 |
| Energy contribution | -28.60 |
| Covariance contribution | -5.15 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.38 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 150318 91 + 5227293/0-120 UCCGCAGUAGCUCAG-UGGUAGAGCUAUCGGCUGUUAACCGAUCGGUCGUAGGUUCGAGUCCUACCUGCGGAGCCAUUAU----------------------------GCUUCCAUAGCU (((((((((((((..-.....))))))..(((((.........)))))(((((.......))))))))))))........----------------------------(((.....))). ( -31.20) >Bcere.0 150324 91 + 5224283/0-120 UCCGCAGUAGCUCAG-UGGUAGAGCUAUCGGCUGUUAACCGAUCGGUCGUAGGUUCGAGUCCUACCUGCGGAGCCAUUAU----------------------------GCUUCCAUAGCU (((((((((((((..-.....))))))..(((((.........)))))(((((.......))))))))))))........----------------------------(((.....))). ( -31.20) >Bhalo.0 267530 94 + 4202352/0-120 UCCACAGUAGCUCAG-UGGUAGAGCAAUCGGCUGUUAACCGAUCGGUCGUAGGUUCGAGUCCUACCUGUGGAGCCAUUUUU-------------------------UUGCUUCCAUAGCU .((((.........)-)))..((.(.(((((.......))))).).))(((((.......)))))((((((((.((.....-------------------------.)).)))))))).. ( -27.90) >Bclau.0 223515 94 + 4303871/0-120 UCCACAGUAGCUCAG-UGGUAGAGCUAUCGGCUGUUAACCGAUCGGUCGCAGGUUCGAAUCCUGCCUGUGGAGCCAUUUUU-------------------------GUGCUUCCAUAGCU ......(((((((..-.....))))))).(((((.........)))))(((((.......)))))(((((((((((....)-------------------------).)).))))))).. ( -32.20) >Bsubt.0 1041786 120 - 4214630/0-120 GGAGGAUUAGCUCAGCUGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCGGCGGUUCGAGCCCGUCAUCCUCCACCAUUUUUCAUUAUACAUAUCGGUUUUACAUAUAUGCCGGUGUAGCU (((((((..(((((((((...((.(.(((((.......))))).).))..))))).)))).....))))))).................((((((((...........)))))))).... ( -39.10) >Blich.0 1104978 103 - 4222334/0-120 GGAGGAUUAGCUCAGCUGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCGGCGGUUCGAUCCCGUCAUCCUCCACCAUU----------CAUAUUGGA-------AUGGGCCGGUGUAGCU (((((((..((((........)))).(((((.......)))))(((((((....)))))))....))))))).(((((----------(......))-------))))............ ( -38.30) >consensus UCCGCAGUAGCUCAG_UGGUAGAGCAAUCGGCUGUUAACCGAUCGGUCGUAGGUUCGAGUCCUACCUGCGGAGCCAUUUU___________________________UGCUUCCAUAGCU (((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))....................................(((.....))). (-33.75 = -28.60 + -5.15)

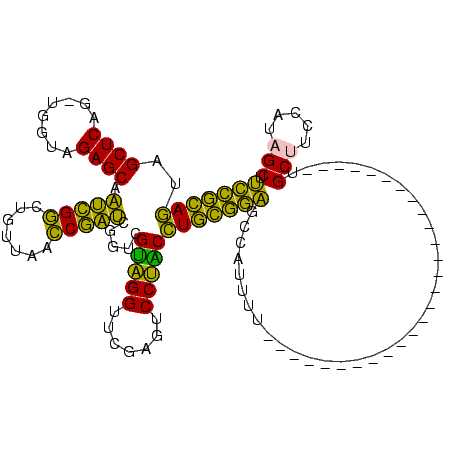
| Location | 150,318 – 150,409 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.58 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -35.68 |
| Energy contribution | -30.75 |
| Covariance contribution | -4.93 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 150318 91 + 5227293/0-120 AGCUAUGGAAGC----------------------------AUAAUGGCUCCGCAGGUAGGACUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUACCA-CUGAGCUACUGCGGA .(((((......----------------------------...)))))((((((((((((.......))))).....((((((.....))))))(((((.....-..))))).))))))) ( -35.70) >Bcere.0 150324 91 + 5224283/0-120 AGCUAUGGAAGC----------------------------AUAAUGGCUCCGCAGGUAGGACUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUACCA-CUGAGCUACUGCGGA .(((((......----------------------------...)))))((((((((((((.......))))).....((((((.....))))))(((((.....-..))))).))))))) ( -35.70) >Bhalo.0 267530 94 + 4202352/0-120 AGCUAUGGAAGCAA-------------------------AAAAAUGGCUCCACAGGUAGGACUCGAACCUACGACCGAUCGGUUAACAGCCGAUUGCUCUACCA-CUGAGCUACUGUGGA .(((((........-------------------------....)))))((((((((((((.......)))))....(((((((.....)))))))((((.....-..))))..))))))) ( -33.80) >Bclau.0 223515 94 + 4303871/0-120 AGCUAUGGAAGCAC-------------------------AAAAAUGGCUCCACAGGCAGGAUUCGAACCUGCGACCGAUCGGUUAACAGCCGAUAGCUCUACCA-CUGAGCUACUGUGGA .(((((........-------------------------....)))))((((((((((((.......))))).....((((((.....))))))(((((.....-..))))).))))))) ( -35.90) >Bsubt.0 1041786 120 - 4214630/0-120 AGCUACACCGGCAUAUAUGUAAAACCGAUAUGUAUAAUGAAAAAUGGUGGAGGAUGACGGGCUCGAACCGCCGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAUCCUCC .....(((((.((((((((((.......))))))).))).....)))))((((((...(((.(((......))))))((((((.....)))))).((((........))))..)))))). ( -37.40) >Blich.0 1104978 103 - 4222334/0-120 AGCUACACCGGCCCAU-------UCCAAUAUG----------AAUGGUGGAGGAUGACGGGAUCGAACCGCCGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAUCCUCC .(((.....)))((((-------((......)----------))))).(((((((...(((.(((......))))))((((((.....)))))).((((........))))..))))))) ( -37.60) >consensus AGCUAUGGAAGCA___________________________AAAAUGGCUCCGCAGGUAGGACUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUACCA_CUGAGCUACUGCGGA .(((.....)))....................................((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))) (-35.68 = -30.75 + -4.93)


| Location | 150,318 – 150,410 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.26 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -25.07 |
| Energy contribution | -21.30 |
| Covariance contribution | -3.77 |
| Combinations/Pair | 1.58 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 150318 92 + 5227293/40-160 GAUCGGUCGUAGGUUCGAGUCCUACCUGCGGAGCCAUUAU----------------------------GCUUCCAUAGCUCAGCUGGUAGAGCACUUCCAUGGUAAGGAAGAGGUCACCG ((((..(((......)))(..(((((.((.((((...(((----------------------------(....)))))))).)).)))))..).(((((.......))))).)))).... ( -30.40) >Bcere.0 150324 92 + 5224283/40-160 GAUCGGUCGUAGGUUCGAGUCCUACCUGCGGAGCCAUUAU----------------------------GCUUCCAUAGCUCAGCUGGUAGAGCACUUCCAUGGUAAGGAAGAGGUCACCG ((((..(((......)))(..(((((.((.((((...(((----------------------------(....)))))))).)).)))))..).(((((.......))))).)))).... ( -30.40) >Bhalo.0 267530 95 + 4202352/40-160 GAUCGGUCGUAGGUUCGAGUCCUACCUGUGGAGCCAUUUUU-------------------------UUGCUUCCAUAGCUCAGUCGGUAGAGCACUACCAUGGUAAGGUAGGGGUCAGCG ............(((.((.((((((((((((((.((.....-------------------------.)).)))))).((.((...(((((....))))).)))).)))))))).))))). ( -31.20) >Bclau.0 223515 95 + 4303871/40-160 GAUCGGUCGCAGGUUCGAAUCCUGCCUGUGGAGCCAUUUUU-------------------------GUGCUUCCAUAGCUCAGUCGGUAGAGCACCACCAUGGUAAGGUGGGGGUCAGCG (((((.(((((((.......)))))(((((((((((....)-------------------------).)).)))))))...........)).).(((((.......))))).)))).... ( -34.50) >Bsubt.0 1041786 120 - 4214630/40-160 AGAGGGUCGGCGGUUCGAGCCCGUCAUCCUCCACCAUUUUUCAUUAUACAUAUCGGUUUUACAUAUAUGCCGGUGUAGCUCAAUUGGUAGAGCAACUGACUUGUAAUCAGUAGGUUGGGG .((((((.(((((.......)))))))))))..((..............((((((((...........)))))))).((((........))))..(..(((((.......)))))..))) ( -34.90) >Blich.0 1104978 103 - 4222334/40-160 AGAGGGUCGGCGGUUCGAUCCCGUCAUCCUCCACCAUU----------CAUAUUGGA-------AUGGGCCGGUGUAGCUCAAUUGGUAGAGCAACUGAAUCGUAAUCAGUAGGUUGGGG .((((((.(((((.......)))))))))))..(((((----------(......))-------))))(((......((((........)))).(((((.......))))).)))..... ( -34.90) >consensus GAUCGGUCGUAGGUUCGAGUCCUACCUGCGGAGCCAUUUU___________________________UGCUUCCAUAGCUCAGUUGGUAGAGCACCUCCAUGGUAAGGAAGAGGUCAGCG ........(((((.......)))))..((...(((..........................................((((........)))).(((((.......))))).)))..)). (-25.07 = -21.30 + -3.77)

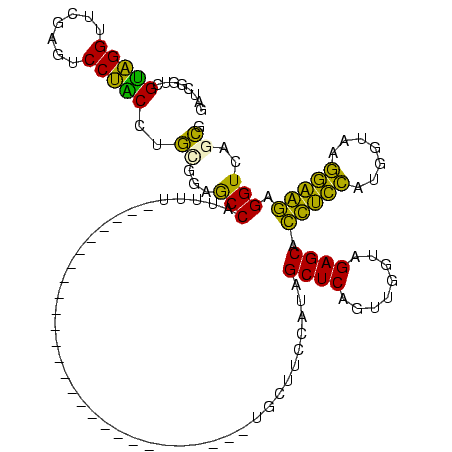
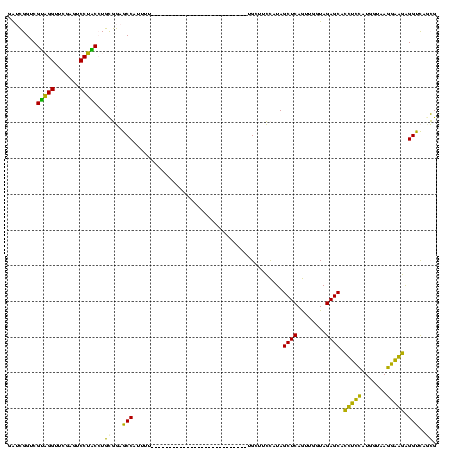
| Location | 150,318 – 150,410 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.26 |
| Mean single sequence MFE | -29.85 |
| Consensus MFE | -21.48 |
| Energy contribution | -17.71 |
| Covariance contribution | -3.77 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 150318 92 + 5227293/40-160 CGGUGACCUCUUCCUUACCAUGGAAGUGCUCUACCAGCUGAGCUAUGGAAGC----------------------------AUAAUGGCUCCGCAGGUAGGACUCGAACCUACGACCGAUC ((((.....(((((.......))))).(..(((((.((.(((((((......----------------------------...))))))).)).)))))..)...........))))... ( -31.10) >Bcere.0 150324 92 + 5224283/40-160 CGGUGACCUCUUCCUUACCAUGGAAGUGCUCUACCAGCUGAGCUAUGGAAGC----------------------------AUAAUGGCUCCGCAGGUAGGACUCGAACCUACGACCGAUC ((((.....(((((.......))))).(..(((((.((.(((((((......----------------------------...))))))).)).)))))..)...........))))... ( -31.10) >Bhalo.0 267530 95 + 4202352/40-160 CGCUGACCCCUACCUUACCAUGGUAGUGCUCUACCGACUGAGCUAUGGAAGCAA-------------------------AAAAAUGGCUCCACAGGUAGGACUCGAACCUACGACCGAUC ...(((..(((((((.....((((((....))))))...(((((((........-------------------------....)))))))...)))))))..)))............... ( -26.70) >Bclau.0 223515 95 + 4303871/40-160 CGCUGACCCCCACCUUACCAUGGUGGUGCUCUACCGACUGAGCUAUGGAAGCAC-------------------------AAAAAUGGCUCCACAGGCAGGAUUCGAACCUGCGACCGAUC ..(((....(((((.......))))).((((........))))..((((.((.(-------------------------(....)))))))))))(((((.......)))))........ ( -29.60) >Bsubt.0 1041786 120 - 4214630/40-160 CCCCAACCUACUGAUUACAAGUCAGUUGCUCUACCAAUUGAGCUACACCGGCAUAUAUGUAAAACCGAUAUGUAUAAUGAAAAAUGGUGGAGGAUGACGGGCUCGAACCGCCGACCCUCU .........((((((.....))))))..((((((((.....(((.....)))((((((((.......)))))))).........))))))))...((.(((.(((......)))))))). ( -31.10) >Blich.0 1104978 103 - 4222334/40-160 CCCCAACCUACUGAUUACGAUUCAGUUGCUCUACCAAUUGAGCUACACCGGCCCAU-------UCCAAUAUG----------AAUGGUGGAGGAUGACGGGAUCGAACCGCCGACCCUCU (((((.((.(((((.......))))).((((........))))......)).((((-------((......)----------)))))))).))..((.(((.(((......)))))))). ( -29.50) >consensus CGCUGACCUCCUCCUUACCAUGGAAGUGCUCUACCAACUGAGCUAUGGAAGCA___________________________AAAAUGGCUCCGCAGGUAGGACUCGAACCUACGACCGAUC .........(((((.......))))).((((........))))..((((.((..................................))))))...(((((.......)))))........ (-21.48 = -17.71 + -3.77)
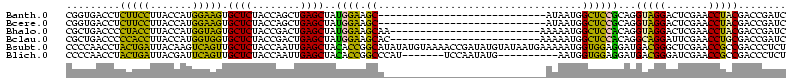
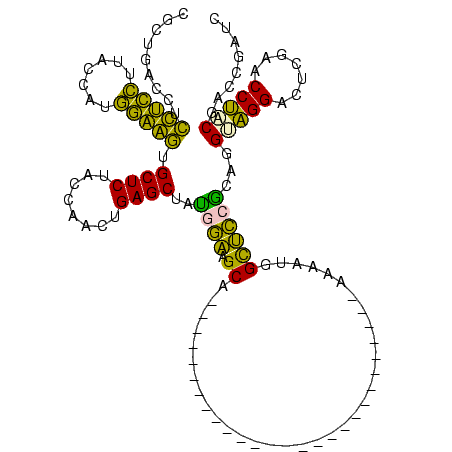
| Location | 150,318 – 150,410 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.51 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -33.22 |
| Energy contribution | -28.07 |
| Covariance contribution | -5.16 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.38 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
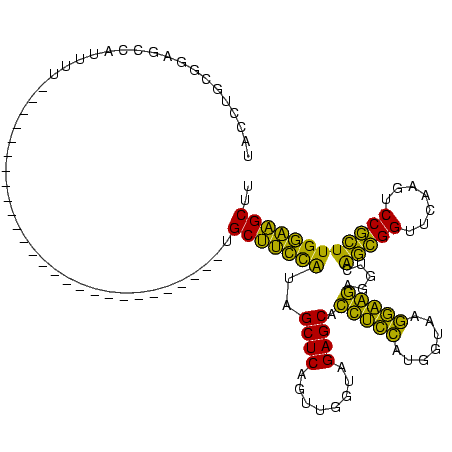
>Banth.0 150318 92 + 5227293/62-182 UACCUGCGGAGCCAUUAU----------------------------GCUUCCAUAGCUCAGCUGGUAGAGCACUUCCAUGGUAAGGAAGAGGUCACCGGUUCAAGCCCGGUUGGAAGCUU ..................----------------------------(((((((..(((.(((((((.((.(.(((((.......))))).).)))))))))..))).....))))))).. ( -31.20) >Bcere.0 150324 92 + 5224283/62-182 UACCUGCGGAGCCAUUAU----------------------------GCUUCCAUAGCUCAGCUGGUAGAGCACUUCCAUGGUAAGGAAGAGGUCACCGGUUCAAGCCCGGUUGGAAGCUU ..................----------------------------(((((((..(((.(((((((.((.(.(((((.......))))).).)))))))))..))).....))))))).. ( -31.20) >Bhalo.0 267530 95 + 4202352/62-182 UACCUGUGGAGCCAUUUUU-------------------------UUGCUUCCAUAGCUCAGUCGGUAGAGCACUACCAUGGUAAGGUAGGGGUCAGCGGUUCAAGUCCGCUUGGAAGCUU .((((((((((.((.....-------------------------.)).)))))).((((........)))).(((((.......))))))))).(((..((((((....)))))).))). ( -30.60) >Bclau.0 223515 95 + 4303871/62-182 UGCCUGUGGAGCCAUUUUU-------------------------GUGCUUCCAUAGCUCAGUCGGUAGAGCACCACCAUGGUAAGGUGGGGGUCAGCGGUUCAAGUCCGCUUGGAAGCUU .(((((((((((((....)-------------------------).)).))))).((((........)))).(((((.......))))))))).(((..((((((....)))))).))). ( -34.40) >Bsubt.0 1041786 120 - 4214630/62-182 GUCAUCCUCCACCAUUUUUCAUUAUACAUAUCGGUUUUACAUAUAUGCCGGUGUAGCUCAAUUGGUAGAGCAACUGACUUGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUUGCCGGCAC (((..((((((((..............((((((((...........)))))))).((((........)))).(((((.......))))).)).))))))..((((....))))..))).. ( -33.60) >Blich.0 1104978 103 - 4222334/62-182 GUCAUCCUCCACCAUU----------CAUAUUGGA-------AUGGGCCGGUGUAGCUCAAUUGGUAGAGCAACUGAAUCGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUUGCCGGCAC ...........(((((----------(......))-------))))((((((...((((........)))).(((((.......))))).....(((((......)))))..)))))).. ( -33.60) >consensus UACCUGCGGAGCCAUUUU___________________________UGCUUCCAUAGCUCAGUUGGUAGAGCACCUCCAUGGUAAGGAAGAGGUCAGCGGUUCAAGUCCGCUUGGAAGCUU ..............................................(((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))).. (-33.22 = -28.07 + -5.16)


| Location | 150,318 – 150,410 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.51 |
| Mean single sequence MFE | -29.91 |
| Consensus MFE | -27.85 |
| Energy contribution | -25.13 |
| Covariance contribution | -2.71 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
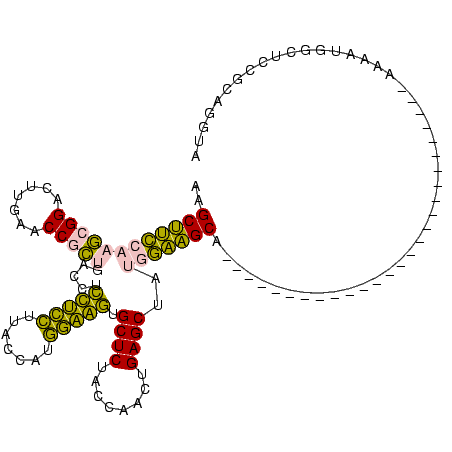
>Banth.0 150318 92 + 5227293/62-182 AAGCUUCCAACCGGGCUUGAACCGGUGACCUCUUCCUUACCAUGGAAGUGCUCUACCAGCUGAGCUAUGGAAGC----------------------------AUAAUGGCUCCGCAGGUA ((((((......))))))..((((((((.(.(((((.......))))).).)).))).((.(((((((......----------------------------...))))))).)).))). ( -31.40) >Bcere.0 150324 92 + 5224283/62-182 AAGCUUCCAACCGGGCUUGAACCGGUGACCUCUUCCUUACCAUGGAAGUGCUCUACCAGCUGAGCUAUGGAAGC----------------------------AUAAUGGCUCCGCAGGUA ((((((......))))))..((((((((.(.(((((.......))))).).)).))).((.(((((((......----------------------------...))))))).)).))). ( -31.40) >Bhalo.0 267530 95 + 4202352/62-182 AAGCUUCCAAGCGGACUUGAACCGCUGACCCCUACCUUACCAUGGUAGUGCUCUACCGACUGAGCUAUGGAAGCAA-------------------------AAAAAUGGCUCCACAGGUA ..((((((((((((.......))))).....(((((.......))))).((((........))))..)))))))..-------------------------................... ( -30.00) >Bclau.0 223515 95 + 4303871/62-182 AAGCUUCCAAGCGGACUUGAACCGCUGACCCCCACCUUACCAUGGUGGUGCUCUACCGACUGAGCUAUGGAAGCAC-------------------------AAAAAUGGCUCCACAGGCA ..((((((((((((.......))))).....(((((.......))))).((((........))))..)))))))..-------------------------.......(((.....))). ( -33.80) >Bsubt.0 1041786 120 - 4214630/62-182 GUGCCGGCAAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUGCUCUACCAAUUGAGCUACACCGGCAUAUAUGUAAAACCGAUAUGUAUAAUGAAAAAUGGUGGAGGAUGAC ..(((((......((((((....................)))))).((.((((........)))).)).)))))((((((((.......))))))))....................... ( -26.45) >Blich.0 1104978 103 - 4222334/62-182 GUGCCGGCAAGAGGACUUGAACCCCCAACCUACUGAUUACGAUUCAGUUGCUCUACCAAUUGAGCUACACCGGCCCAU-------UCCAAUAUG----------AAUGGUGGAGGAUGAC ..(((((((((....))))............(((((.......))))).((((........))))....)))))((((-------((......)----------)))))........... ( -26.40) >consensus AAGCUUCCAAGCGGACUUGAACCGCUGACCUCCUCCUUACCAUGGAAGUGCUCUACCAACUGAGCUAUGGAAGCA___________________________AAAAUGGCUCCGCAGGUA ..((((((((((((.......))))).....(((((.......))))).((((........))))..))))))).............................................. (-27.85 = -25.13 + -2.71)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:19:23 2006