
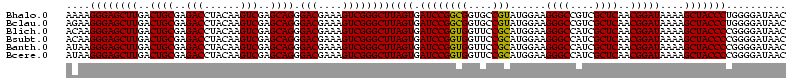
| Sequence ID | Bhalo.0 |
|---|---|
| Location | 3,178,984 – 3,179,104 |
| Length | 120 |
| Max. P | 0.999994 |

| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.08 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -23.37 |
| Energy contribution | -20.77 |
| Covariance contribution | -2.60 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
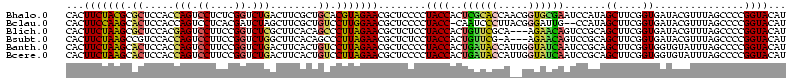
>Bhalo.0 3178984 120 - 4202352/40-160 CCCUUAGAGAUGAUGAGGUUGAUAGGUCUGGUGUGGAAGCGUGGCGACACGUGAAGCUGACAGAUACUAAUCGGUCGAGGACUUAUCCAAAAACAAAUCAAAAGCAACGUCUCGAACUCG ......((((((.((.(..(((((((((((..((....(((((....)))))...))...))))).)).))))..)..(((....)))................)).))))))....... ( -33.20) >Bclau.0 2858068 120 - 4303871/40-160 CCCUUAGAGAUGAUGAGGUUGAUAGGUCAGAAGUGGAAGCGUGGCGACACGUGGAGCGGACUGAUACUAAUCGGUCGAGGACUUAUCCUAUAUACAUUCUUGACAAUUGAGUCUAUUUCG ......((((((((..((((((((((((....((....(((((....)))))...)).(((((((....)))))))...)))))))........((....)).)))))..))).))))). ( -34.80) >Blich.0 3118231 115 + 4222334/40-160 CCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAUACUAAUCGAUCGAGGACUUAACCUAAAUAUUUGUGAAUCGUCU---GUCCGUU-- ....((.(((((((((((((((((((((((.(((....(((((....)))))...)))..))))).)).))))))).(((......))).........)).)))))))---.))....-- ( -33.90) >Bsubt.0 3172985 115 + 4214630/40-160 CCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCAACACAUGGAGCUGACAGAUACUAAUCGAUCGAGGACUUAACC---AUAUUUUUGAAUGAUGUCACACCUGUU-- ..............((((((((((.(((((.(((....(((((....)))))...)))..))))).....(((.(((((((.......---...))))))).)))))))).)))))..-- ( -30.60) >Banth.0 5077122 108 - 5227293/40-160 CCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC-AUAUAAUAUGAAGCGAU---------GUU-- .(((((.......)))))(((((..(((((.(((....(((((....)))))...)))..)))))........)))))((......))-.................---------...-- ( -23.70) >Bcere.0 5074106 108 - 5224283/40-160 CCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC-AUAUAAUAUGUAGCAAU---------GUU-- .(((((.......)))))(((((..(((((.(((....(((((....)))))...)))..)))))........)))))((......))-....(((((......))---------)))-- ( -23.80) >consensus CCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGCGUGGCGACACGUGGAGCUGACAGAUACUAAUCGAUCGAGGACUUAACC_AUAUAAUAUCAAAAGAU_U____C__GUU__ .(((((.......)))))(((((..(((((..((....(((((....)))))...))...)))))........)))))((......))................................ (-23.37 = -20.77 + -2.60)
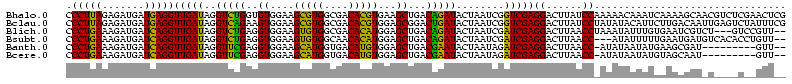
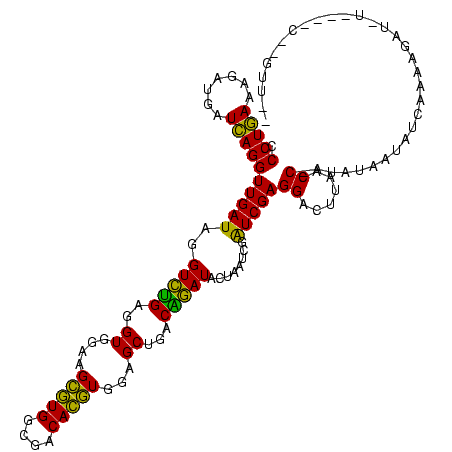
| Location | 3,178,984 – 3,179,103 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.36 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -32.25 |
| Energy contribution | -28.32 |
| Covariance contribution | -3.94 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 119 - 4202352/80-200 CUCAAGAUGAGAUUUCCCAUGGA-GUAAAUCCAGUAAGACCCCUUAGAGAUGAUGAGGUUGAUAGGUCUGGUGUGGAAGCGUGGCGACACGUGAAGCUGACAGAUACUAAUCGGUCGAGG (((................((((-.....))))....(((((((((.......)))))..((((((((((..((....(((((....)))))...))...))))).)).)))))))))). ( -35.30) >Bclau.0 2858068 118 - 4303871/80-200 CUCAAGAUGAGAUUUCCCAUGGC-GUAAG-CCAGUAAGACCCCUUAGAGAUGAUGAGGUUGAUAGGUCAGAAGUGGAAGCGUGGCGACACGUGGAGCGGACUGAUACUAAUCGGUCGAGG (((..........(((((.((((-....)-)))....(((((((((.......)))))......))))....).))))(((((....)))))......(((((((....)))))))))). ( -39.20) >Blich.0 3118231 119 + 4222334/80-200 CUCAAGAUGAGAUUUCCCAUUCCUAUAAG-GAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAUACUAAUCGAUCGAGG (((..(((.(..((((...((((.....)-)))...((....))))))..).))).((((((((((((((.(((....(((((....)))))...)))..))))).)).)))))))))). ( -34.20) >Bsubt.0 3172985 118 + 4214630/80-200 CUCAAGAUGAGAUUUCCCAUUCC-GCAAG-GAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCAACACAUGGAGCUGACAGAUACUAAUCGAUCGAGG (((..(((.(..((((...((((-....)-)))...((....))))))..).))).((((((((((((((.(((....(((((....)))))...)))..))))).)).)))))))))). ( -38.70) >Banth.0 5077122 118 - 5227293/80-200 CUCAAGAUGAGAUUUCCCAUAGC-GUAAG-CUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGG (((................((((-....)-)))....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))))). ( -35.90) >Bcere.0 5074106 118 - 5224283/80-200 CUCAAGAUGAGAUUUCCCAUAGC-GUAAG-CUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGG (((................((((-....)-)))....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))))). ( -35.90) >consensus CUCAAGAUGAGAUUUCCCAUAGC_GUAAG_CAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGCGUGGCGACACGUGGAGCUGACAGAUACUAAUCGAUCGAGG (((................(((........)))....(((((((((.......)))))....((((((((..((....(((((....)))))...))...))))).)))...))))))). (-32.25 = -28.32 + -3.94)
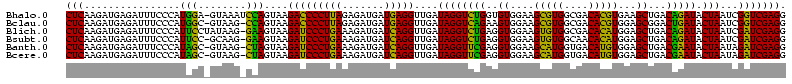
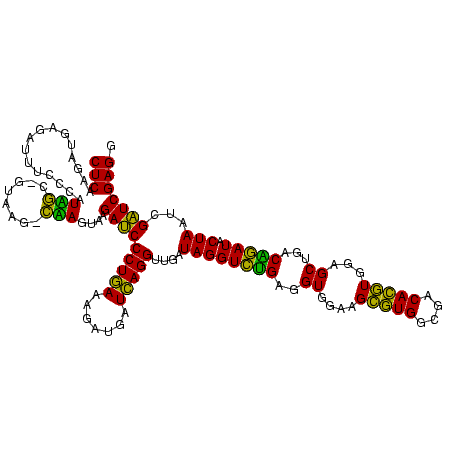
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -45.10 |
| Consensus MFE | -45.87 |
| Energy contribution | -44.43 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -0.80 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/240-360 CUAUCCGUCGCGGGCGUAGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACACACCGCUGGUGUACCAGUUGUUCCGCCAGGAGCAUCGCUGGGUAGCUACGUGUG ...((((((.(((((..((((..(((....)))....))))..)).(....)..))).))))))(((((.(.((((...(((((((((((....))))))..)))))...)))))))))) ( -47.30) >Bclau.0 2858068 120 - 4303871/240-360 CUAUCCGUCGCGGGCGUAGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACAUACCGCUGGUGUACCAGUUGUUCCGCCAGGAGCAUCGCUGGGUAGCUACGUAUG ...((((((.(((((..((((..(((....)))....))))..)).(....)..))).))))))(((((.(.((((...(((((((((((....))))))..)))))...)))))))))) ( -45.40) >Blich.0 3118231 120 + 4222334/240-360 CUAUCCGUCGCGGGCGCAGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACGCACCGCUGGUGUACCAGUUGUUCUGCCAAGGGCAUCGCUGGGUAGCUAUGUGCG ...((((((.(((((..((((..(((....)))....))))..)).(....)..))).))))))(((((...((((...(((((((((((....))))))..)))))...)))).))))) ( -45.40) >Bsubt.0 3172985 120 + 4214630/240-360 CUAUCCGUCGCGGGCGCUGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACGCACCGCUGGUGUACCAGUUGUUCUGCCAAGGGCAUCGCUGGGUAGCUAUGUGCG ...((((((.(((((...(((..(((....)))....)))...)).(....)..))).))))))(((((...((((...(((((((((((....))))))..)))))...)))).))))) ( -45.10) >Banth.0 5077122 120 - 5227293/240-360 CUAUCCGUCGUGGGCGUAGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACGCACCGCUGGUGUACCAGUUGUUCUGCCAAGGGCAUAGCUGGGUAGCUAUGUGCG ...((((((.(((((..((((..(((....)))....))))..)).(....)..))).))))))(((((...((((...(((((((((((....))))))..)))))...)))).))))) ( -43.70) >Bcere.0 5074106 120 - 5224283/240-360 CUAUCCGUCGUGGGCGUAGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACGCACCGCUGGUGUACCAGUUGUUCUGCCAAGGGCAUAGCUGGGUAGCUAUGUGCG ...((((((.(((((..((((..(((....)))....))))..)).(....)..))).))))))(((((...((((...(((((((((((....))))))..)))))...)))).))))) ( -43.70) >consensus CUAUCCGUCGCGGGCGUAGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACGCACCGCUGGUGUACCAGUUGUUCUGCCAAGGGCAUCGCUGGGUAGCUAUGUGCG ...((((((.(((((..((((..(((....)))....))))..)).(....)..))).))))))(((((...((((...(((((((((((....))))))..)))))...)))).))))) (-45.87 = -44.43 + -1.44)

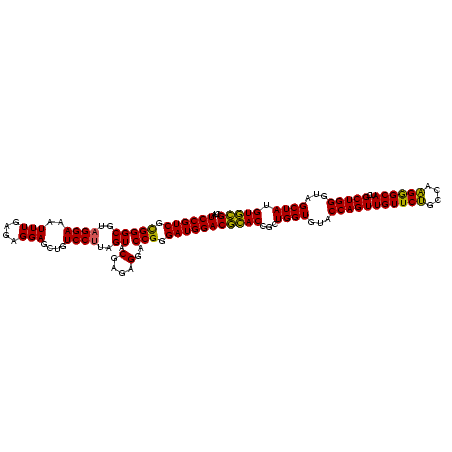

| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.11 |
| Mean single sequence MFE | -47.95 |
| Consensus MFE | -46.58 |
| Energy contribution | -46.38 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/280-400 ACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGUAGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACACACCGCUGGUGUAC ((((.(((.(.((((.(((((((....))).)))).(((((..(((.((.((..(..((((..(((....)))....))))..)..)).)))))..)))))))))...).))).)))).. ( -47.30) >Bclau.0 2858068 120 - 4303871/280-400 ACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGUAGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACAUACCGCUGGUGUAC ((((.(((.(.((((.(((((((....))).)))).(((((..(((.((.((..(..((((..(((....)))....))))..)..)).)))))..)))))))))...).))).)))).. ( -47.30) >Blich.0 3118231 120 + 4222334/280-400 ACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGCAGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACGCACCGCUGGUGUAC ((((.(((((.((((.(((((((....))).)))).(((((..(((.((.((..(..((((..(((....)))....))))..)..)).)))))..))))))))).))..))).)))).. ( -48.70) >Bsubt.0 3172985 120 + 4214630/280-400 ACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGCUGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACGCACCGCUGGUGUAC ((((.(((((.((((.(((((((....))).)))).(((((..(((.((.((..(...(((..(((....)))....)))...)..)).)))))..))))))))).))..))).)))).. ( -48.40) >Banth.0 5077122 120 - 5227293/280-400 ACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGUGGGCGUAGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACGCACCGCUGGUGUAC ((((.(((((.((((.(((((((....))).)))).(((((..(((.(((((.....((((..(((....)))....))))...)))))..)))..))))))))).))..))).)))).. ( -48.00) >Bcere.0 5074106 120 - 5224283/280-400 ACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGUGGGCGUAGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACGCACCGCUGGUGUAC ((((.(((((.((((.(((((((....))).)))).(((((..(((.(((((.....((((..(((....)))....))))...)))))..)))..))))))))).))..))).)))).. ( -48.00) >consensus ACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGUAGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACGCACCGCUGGUGUAC ((((.(((((.((((.(((((((....))).)))).(((((..(((.((.((..(..((((..(((....)))....))))..)..)).)))))..))))))))).))..))).)))).. (-46.58 = -46.38 + -0.19)

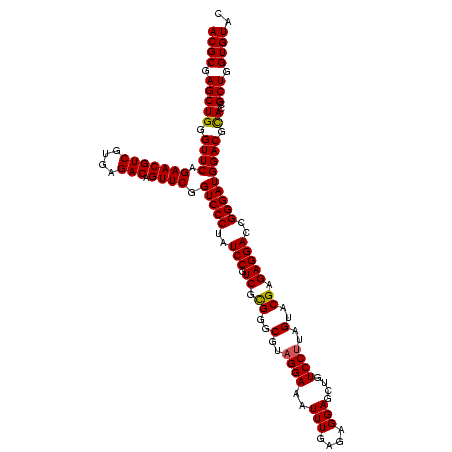
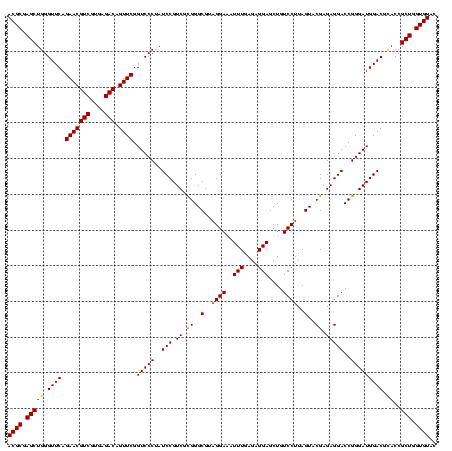
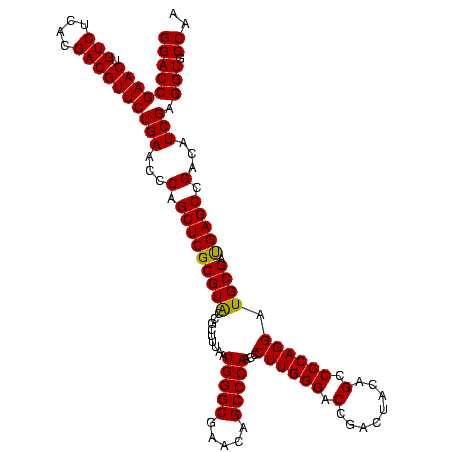
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.56 |
| Mean single sequence MFE | -40.37 |
| Consensus MFE | -40.58 |
| Energy contribution | -40.30 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.76 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/360-480 GGACCGAACUGUCUCACGACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCUACUUCAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAA (((((((((.(((....)))))))(((...(.(((((((((.........(((((.....)))))...((((((.(........).)))))).)))).))))).)...))).))).)).. ( -40.20) >Bclau.0 2858068 120 - 4303871/360-480 GGACCGAACUGUCUCACGACGUUCUGAACCCAGCUCGCGUGCCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCUACUUCAGCCCCAGGAUGCGACGAGCCGACAUCGAGGUGCCAA (((((((((.(((....)))))))(((...(.((((((((..........(((((.....)))))...((((((.(........).))))))..))).))))).)...))).))).)).. ( -41.20) >Blich.0 3118231 120 + 4222334/360-480 GGACCGAACUGUCUCACGACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAA (((((((((.(((....)))))))(((...(.(((((((((.........(((((.....)))))...((((((.(........).)))))).)))).))))).)...))).))).)).. ( -40.20) >Bsubt.0 3172985 120 + 4214630/360-480 GGACCGAACUGUCUCACGACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAA (((((((((.(((....)))))))(((...(.(((((((((.........(((((.....)))))...((((((.(........).)))))).)))).))))).)...))).))).)).. ( -40.20) >Banth.0 5077122 120 - 5227293/360-480 GGACCGAACUGUCUCACGACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAA (((((((((.(((....)))))))(((...(.(((((((((.........(((((.....)))))...((((((.(........).)))))).)))).))))).)...))).))).)).. ( -40.20) >Bcere.0 5074106 120 - 5224283/360-480 GGACCGAACUGUCUCACGACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAA (((((((((.(((....)))))))(((...(.(((((((((.........(((((.....)))))...((((((.(........).)))))).)))).))))).)...))).))).)).. ( -40.20) >consensus GGACCGAACUGUCUCACGACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAA (((((((((.(((....)))))))(((...(.(((((((((.........(((((.....)))))...((((((.(........).)))))).)))).))))).)...))).))).)).. (-40.58 = -40.30 + -0.28)

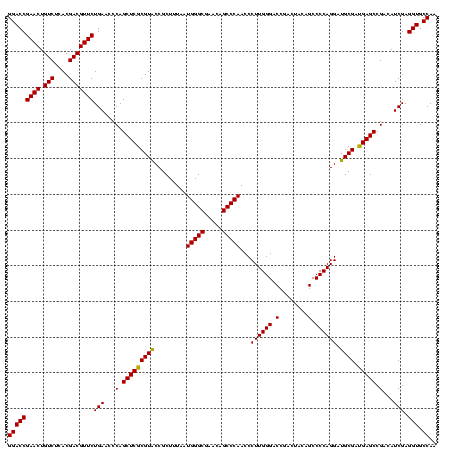
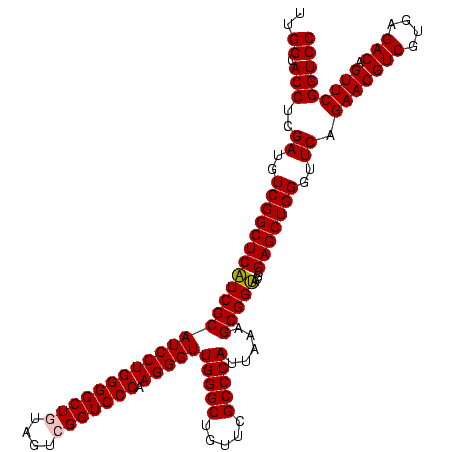
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.56 |
| Mean single sequence MFE | -56.10 |
| Consensus MFE | -55.64 |
| Energy contribution | -55.70 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.19 |
| SVM RNA-class probability | 0.999978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/360-480 UUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGAAGUAGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCC ..((.(((..((..(((((((((((((((((((((((......)))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).))))))))) ( -54.50) >Bclau.0 2858068 120 - 4303871/360-480 UUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGAAGUAGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCC ..((.(((..((..(((((((((((((((((((((((......)))))).)))))(((((.....))))).....)))....)))))))))..)).(((((((....))).))))))))) ( -56.50) >Blich.0 3118231 120 + 4222334/360-480 UUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCC ..((.(((..((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).))))))))) ( -56.40) >Bsubt.0 3172985 120 + 4214630/360-480 UUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCC ..((.(((..((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).))))))))) ( -56.40) >Banth.0 5077122 120 - 5227293/360-480 UUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCC ..((.(((..((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).))))))))) ( -56.40) >Bcere.0 5074106 120 - 5224283/360-480 UUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCC ..((.(((..((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).))))))))) ( -56.40) >consensus UUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCC ..((.(((..((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).))))))))) (-55.64 = -55.70 + 0.06)

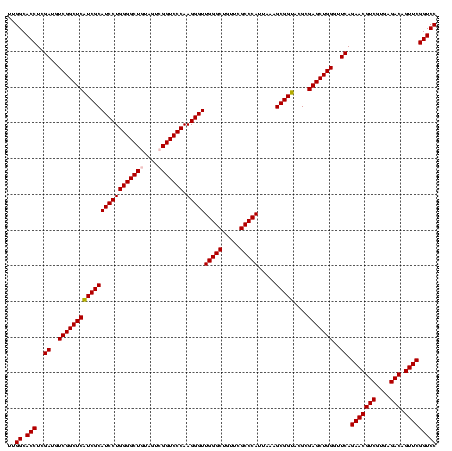
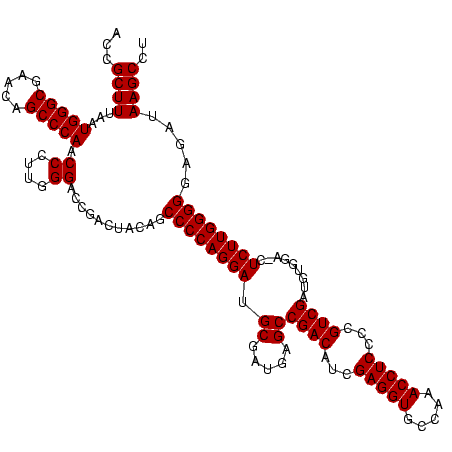
| Location | 3,178,984 – 3,179,103 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.99 |
| Mean single sequence MFE | -42.62 |
| Consensus MFE | -41.00 |
| Energy contribution | -41.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 119 - 4202352/400-520 ACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCUACUUCAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGAA-CUCUUGGGGGAGAUAAGCCU ...((((...(((((.....))))).((....)).......((..((((((((.((.....))((((...(((((.....)))))...)))).......-.))))))))..)).)))).. ( -41.20) >Bclau.0 2858068 119 - 4303871/400-520 GCCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCUACUUCAGCCCCAGGAUGCGACGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGGA-CUCUUGGGGGAGAUAAGCCU ...((((...(((((.....))))).((((..(((.((((.((.((.....((((((......)).))))(((((.....)))))...)).)).)))).-.)))..))))....)))).. ( -43.50) >Blich.0 3118231 119 + 4222334/400-520 ACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGGA-CUCUUGGGGGAGAUAAGCCU ...((((...(((((.....))))).((((..(((....(((((..........((.....))((((...(((((.....)))))...)))).))))).-.)))..))))....)))).. ( -42.40) >Bsubt.0 3172985 120 + 4214630/400-520 ACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGGAGCUCUUGGGGGAGAUAAGCCU ...((((...(((((.....))))).((((..(((.(..(((((..........((.....))((((...(((((.....)))))...)))).))))).).)))..))))....)))).. ( -43.80) >Banth.0 5077122 119 - 5227293/400-520 ACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGGA-CUCUUGGGGGAGAUAAGCCU ...((((...(((((.....))))).((((..(((....(((((..........((.....))((((...(((((.....)))))...)))).))))).-.)))..))))....)))).. ( -42.40) >Bcere.0 5074106 119 - 5224283/400-520 ACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGGA-CUCUUGGGGGAGAUAAGCCU ...((((...(((((.....))))).((((..(((....(((((..........((.....))((((...(((((.....)))))...)))).))))).-.)))..))))....)))).. ( -42.40) >consensus ACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGGA_CUCUUGGGGGAGAUAAGCCU ...((((...(((((.....))))).((....))...........((((((((.((.....))((((...(((((.....)))))...)))).........)))))))).....)))).. (-41.00 = -41.00 + 0.00)

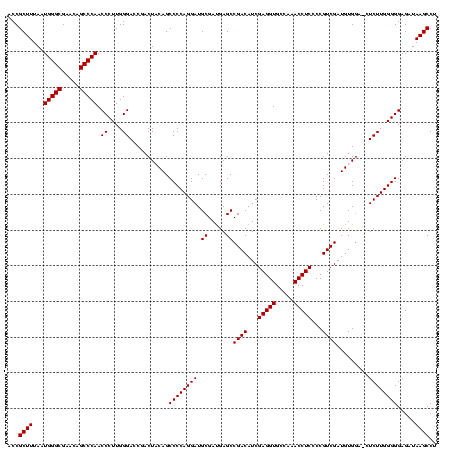
| Location | 3,178,984 – 3,179,103 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.99 |
| Mean single sequence MFE | -51.13 |
| Consensus MFE | -50.29 |
| Energy contribution | -50.65 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.84 |
| SVM RNA-class probability | 0.999656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 119 - 4202352/400-520 AGGCUUAUCUCCCCCAAGAG-UUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGAAGUAGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGU ..(((.((((((((...((.-......))...))))))))..)))(((..((((......))))(((((((((((((......)))))).)))))(((((.....))))).....))))) ( -49.90) >Bclau.0 2858068 119 - 4303871/400-520 AGGCUUAUCUCCCCCAAGAG-UCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGAAGUAGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGC .................(((-((.(((((((.((............))))))))).)))))((((((((((((((((......)))))).)))))(((((.....))))).....))))) ( -49.40) >Blich.0 3118231 119 + 4222334/400-520 AGGCUUAUCUCCCCCAAGAG-UCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGU ..(((.((((((((...((.-......))...))))))))..)))(((..((((......))))((((((((((((((....))))))).)))))(((((.....))))).....))))) ( -51.80) >Bsubt.0 3172985 120 + 4214630/400-520 AGGCUUAUCUCCCCCAAGAGCUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGU ..(((.((((((((...((........))...))))))))..)))(((..((((......))))((((((((((((((....))))))).)))))(((((.....))))).....))))) ( -52.10) >Banth.0 5077122 119 - 5227293/400-520 AGGCUUAUCUCCCCCAAGAG-UCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGU ..(((.((((((((...((.-......))...))))))))..)))(((..((((......))))((((((((((((((....))))))).)))))(((((.....))))).....))))) ( -51.80) >Bcere.0 5074106 119 - 5224283/400-520 AGGCUUAUCUCCCCCAAGAG-UCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGU ..(((.((((((((...((.-......))...))))))))..)))(((..((((......))))((((((((((((((....))))))).)))))(((((.....))))).....))))) ( -51.80) >consensus AGGCUUAUCUCCCCCAAGAG_UCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGU ..(((.((((((((...((........))...))))))))..)))(((..((((......))))((((((((((((((....))))))).)))))(((((.....))))).....))))) (-50.29 = -50.65 + 0.36)

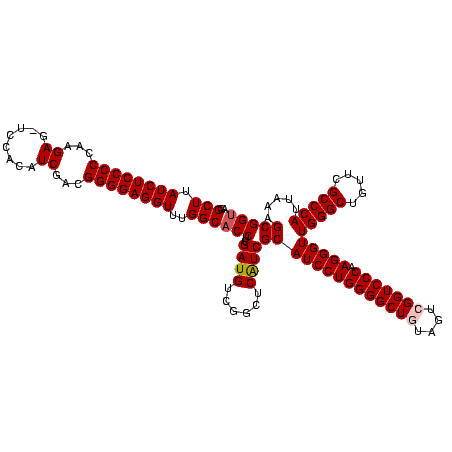
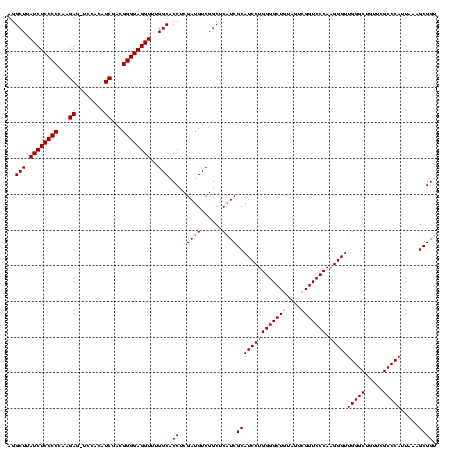
| Location | 3,178,984 – 3,179,103 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.82 |
| Mean single sequence MFE | -42.55 |
| Consensus MFE | -42.13 |
| Energy contribution | -42.10 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.62 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 119 - 4202352/440-560 GGGCCGUCGCUCAACGGAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCAAGAG-UUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGAA ((((....))))............(..(((..(((.......(((.((((((((...((.-......))...))))))))..)))....(((((......)))))..)))..)))..).. ( -39.70) >Bclau.0 2858068 119 - 4303871/440-560 GGGCCGUCGCUCAACGGAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCAAGAG-UCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGAA ((((....))))............(..(((..(((.......(((.((((((((...((.-......))...))))))))..)))....(((((......)))))..)))..)))..).. ( -39.30) >Blich.0 3118231 119 + 4222334/440-560 GGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAG-UCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUA ((((....))))...........((..((((((((.......(((.((((((((...((.-......))...))))))))..)))....(((((......)))))..))))))))..)). ( -44.00) >Bsubt.0 3172985 120 + 4214630/440-560 GGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGCUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUA ((((....))))...........((..((((((((.......(((.((((((((...((........))...))))))))..)))....(((((......)))))..))))))))..)). ( -44.30) >Banth.0 5077122 119 - 5227293/440-560 GGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAG-UCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUA ((((....))))...........((..((((((((.......(((.((((((((...((.-......))...))))))))..)))....(((((......)))))..))))))))..)). ( -44.00) >Bcere.0 5074106 119 - 5224283/440-560 GGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAG-UCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUA ((((....))))...........((..((((((((.......(((.((((((((...((.-......))...))))))))..)))....(((((......)))))..))))))))..)). ( -44.00) >consensus GGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAG_UCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUA ((((....))))...........((..((((((((.......(((.((((((((...((........))...))))))))..)))....(((((......)))))..))))))))..)). (-42.13 = -42.10 + -0.03)

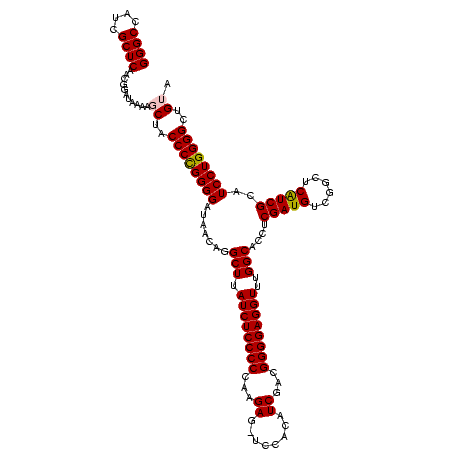
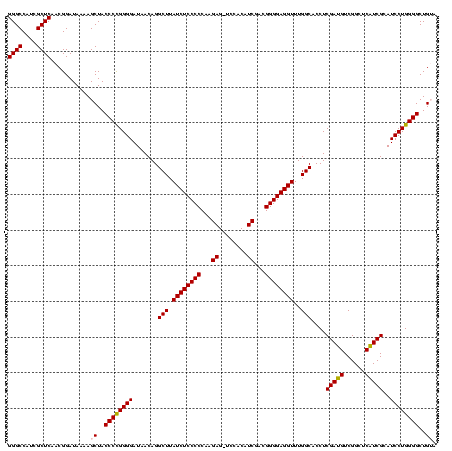
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -43.10 |
| Consensus MFE | -42.44 |
| Energy contribution | -42.00 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/520-640 AAAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGCGGUGCCGUAUGGAAGGGCCGUCGCUCAACGGAUAAAAGCUACCCUGGGGAUAAC ...(((((((((..((((..(((......)))..)))).(((....))))))))((((.((((((((((.((........))))))))(.....)))))....))))))))......... ( -43.50) >Bclau.0 2858068 120 - 4303871/520-640 AGAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGCGGUGCCGUAUGGAAGGGCCGUCGCUCAACGGAUAAAAGCUACCCUGGGGAUAAC ...(((((((((..((((..(((......)))..)))).(((....))))))))((((.((((((((((.((........))))))))(.....)))))....))))))))......... ( -43.50) >Blich.0 3118231 120 + 4222334/520-640 ACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAAC .(..((((((((..((((..(((......)))..)))).(((....))))))))((((.((((((((....)))......((((....))))..)))))....)))).)))..)...... ( -43.20) >Bsubt.0 3172985 120 + 4214630/520-640 ACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAAC .(..((((((((..((((..(((......)))..)))).(((....))))))))((((.((((((((....)))......((((....))))..)))))....)))).)))..)...... ( -43.20) >Banth.0 5077122 120 - 5227293/520-640 AUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAAC ....((((((((..((((..(((......)))..)))).(((....))))))))((((.((((((((....)))......((((....))))..)))))....)))).)))......... ( -42.60) >Bcere.0 5074106 120 - 5224283/520-640 AUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAAC ....((((((((..((((..(((......)))..)))).(((....))))))))((((.((((((((....)))......((((....))))..)))))....)))).)))......... ( -42.60) >consensus ACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAAC ....((((((((..((((..(((......)))..)))).(((....))))))))((((.((((((((....)))......((((....))))..)))))....))))))).......... (-42.44 = -42.00 + -0.44)
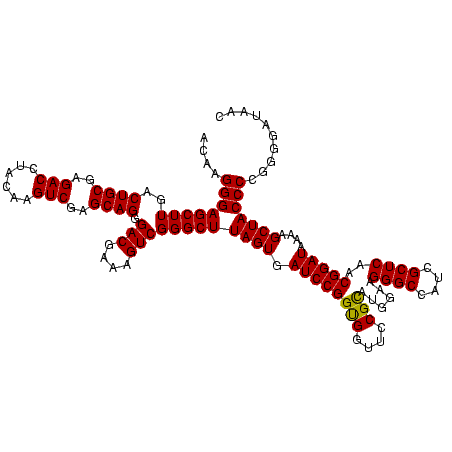
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -41.30 |
| Consensus MFE | -41.31 |
| Energy contribution | -40.87 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.85 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/560-680 CCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAAAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGCGGUGCCGUAUGGAA ((((..(((((((.....)))))))......(((....)))..))))(((((..((((..(((......)))..)))).(((....))))))))......(((((((....))).)))). ( -39.70) >Bclau.0 2858068 120 - 4303871/560-680 CCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAGAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGCGGUGCCGUAUGGAA (((((.(((((((.....)))))))......(((....)))).))))(((((..((((..(((......)))..)))).(((....))))))))......(((((((....))).)))). ( -40.30) >Blich.0 3118231 120 + 4222334/560-680 CCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAA ((((..(((((((.....))))))).....((((....)))).))))(((((..((((..(((......)))..)))).(((....))))))))......(((((((....))).)))). ( -42.20) >Bsubt.0 3172985 120 + 4214630/560-680 CCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAA ((((..(((((((.....))))))).....((((....)))).))))(((((..((((..(((......)))..)))).(((....))))))))......(((((((....))).)))). ( -42.20) >Banth.0 5077122 120 - 5227293/560-680 CCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAA ((((..(((((((.....))))))).....((((....)))).))))(((((..((((..(((......)))..)))).(((....))))))))......(((((((....))).)))). ( -41.70) >Bcere.0 5074106 120 - 5224283/560-680 CCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAA ((((..(((((((.....))))))).....((((....)))).))))(((((..((((..(((......)))..)))).(((....))))))))......(((((((....))).)))). ( -41.70) >consensus CCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAA ((((..(((((((.....))))))).....((((....)))).))))(((((..((((..(((......)))..)))).(((....))))))))......(((((((....))).)))). (-41.31 = -40.87 + -0.44)
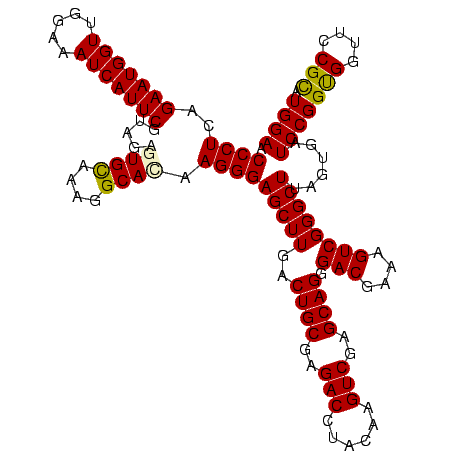
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -39.18 |
| Consensus MFE | -37.08 |
| Energy contribution | -37.42 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.998474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/600-720 CCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUUUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACUGUUUAGGAGGCGACCGC ..((((..((((....))))..)))).((((.((((((.(((....)))......(((((.........))))).))))))...))))(((((((((...........))))))).)).. ( -38.40) >Bclau.0 2858068 120 - 4303871/600-720 CCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUCUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACUGUUUAGGAGGCGACCGC ..((((..((((....))))..)))).((((.((((((.(((....)))......(((((.........))))).))))))...))))(((((((((...........))))))).)).. ( -38.30) >Blich.0 3118231 120 + 4222334/600-720 CCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUGUGCCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGC ..((((..((((....))))..)))).((((.(((((((((......))).....(((((.........))))).))))))...))))(((((((((...........))))))).)).. ( -39.30) >Bsubt.0 3172985 120 + 4214630/600-720 CCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUGUGCCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGC ..((((..((((....))))..)))).((((.(((((((((......))).....(((((.........))))).))))))...))))(((((((((...........))))))).)).. ( -39.30) >Banth.0 5077122 120 - 5227293/600-720 CCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGC ..((((..((((....))))..)))).((((.((((((((((....)))......(((((.........))))))))))))...))))(((((((((...........))))))).)).. ( -39.90) >Bcere.0 5074106 120 - 5224283/600-720 CCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGC ..((((..((((....))))..)))).((((.((((((((((....)))......(((((.........))))))))))))...))))(((((((((...........))))))).)).. ( -39.90) >consensus CCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGC ..((((..((((....))))..)))).((((.((((((.(((....)))......(((((.........))))).))))))...))))(((((((((...........))))))).)).. (-37.08 = -37.42 + 0.33)
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -48.60 |
| Consensus MFE | -48.17 |
| Energy contribution | -48.17 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.85 |
| SVM RNA-class probability | 0.999994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/600-720 GCGGUCGCCUCCUAAACAGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAAAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGG ..((.(((((((...........))))))).))..(((((((((..(((((((.....)))))))......(((....)))..)))))))))..((((..(((......)))..)))).. ( -47.40) >Bclau.0 2858068 120 - 4303871/600-720 GCGGUCGCCUCCUAAACAGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAGAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGG ..((.(((((((...........))))))).))..((((((((((.(((((((.....)))))))......(((....)))).)))))))))..((((..(((......)))..)))).. ( -48.00) >Blich.0 3118231 120 + 4222334/600-720 GCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGG ..((.(((((((...........))))))).))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).. ( -49.30) >Bsubt.0 3172985 120 + 4214630/600-720 GCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGG ..((.(((((((...........))))))).))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).. ( -49.30) >Banth.0 5077122 120 - 5227293/600-720 GCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGG ..((.(((((((...........))))))).))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).. ( -48.80) >Bcere.0 5074106 120 - 5224283/600-720 GCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGG ..((.(((((((...........))))))).))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).. ( -48.80) >consensus GCGGUCGCCUCCUAAAAAGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGG ..((.(((((((...........))))))).))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).. (-48.17 = -48.17 + 0.00)
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.28 |
| Mean single sequence MFE | -49.52 |
| Consensus MFE | -46.81 |
| Energy contribution | -43.18 |
| Covariance contribution | -3.63 |
| Combinations/Pair | 1.39 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/680-800 AACCUUUGGGCGCCUCCGUUACUGUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCCCUGAACCGGAUCACGGUUCGAGGUUAGAAUGUCAAUACCGUCAGGGUAGU ......(((((((((((...........))))))(((.......))).....)))))((((((.(((.((.(((((((.....))))))).)).))).((....))...))))))..... ( -47.10) >Bclau.0 2858068 120 - 4303871/680-800 AACCUUUGGGCGCCUCCGUUACUGUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACAAUGUCCCUGGCCCGGAUCACGGGUCGAGGUUAGAAUGCCAGCACCAUCAGGGUAGU ......(((((((((((...........))))))(((.......))).....)))))(((((...(((...(((((((.....))))))).(((......))).)))...)))))..... ( -46.10) >Blich.0 3118231 120 + 4222334/680-800 AACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCUGCCCGAUAAGGGCAGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGU .....((((((((((((...........))))))).....)))))....(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).))))))) ( -49.40) >Bsubt.0 3172985 120 + 4214630/680-800 AACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCCGCCCGAUAAGGGCGGGGGGUUAGAAGGUCAAUACAGCCAGGGUAGU .((((...(((((((((...........))))))((((......((((...........)))).(((.((((((((((.....)))))))))).)))..))))......))).))))... ( -57.70) >Banth.0 5077122 120 - 5227293/680-800 AACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGU .(((..(((((((((((...........))))))(((.......))).....)))))((((((.(((.((..((((((.....))))))..)).))).((....))...))))))..))) ( -48.40) >Bcere.0 5074106 120 - 5224283/680-800 AACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGU .(((..(((((((((((...........))))))(((.......))).....)))))((((((.(((.((..((((((.....))))))..)).))).((....))...))))))..))) ( -48.40) >consensus AACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGCCCGAUAAGGGCCGGAGGUUAGAAUGUCAAUACAGUCAGGGUAGU .....((((((((((((...........))))))).....)))))....((((((((((((((...)))...((((((.....)))))).))))..(.....)..........))))))) (-46.81 = -43.18 + -3.63)

| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.28 |
| Mean single sequence MFE | -49.60 |
| Consensus MFE | -48.79 |
| Energy contribution | -44.97 |
| Covariance contribution | -3.83 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/680-800 ACUACCCUGACGGUAUUGACAUUCUAACCUCGAACCGUGAUCCGGUUCAGGGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAACAGUAACGGAGGCGCCCAAAGGUU ..((((.....))))((((((((....(((.((((((.....)))))))))...)))))))).(((((.....(((((...)))))((((((...........)))))))))))...... ( -47.70) >Bclau.0 2858068 120 - 4303871/680-800 ACUACCCUGAUGGUGCUGGCAUUCUAACCUCGACCCGUGAUCCGGGCCAGGGACAUUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAACAGUAACGGAGGCGCCCAAAGGUU .....(((((..((((((((..((.......)).(((.....))))))))...)))..)))))(((((.....(((((...)))))((((((...........)))))))))))...... ( -47.80) >Blich.0 3118231 120 + 4222334/680-800 ACUACCCUGGCUGUAUUGACCUUCUAACCCGCUGCCCUUAUCGGGCAGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUU ....(((.((((((.((((((((((...((.((((((.....))))))))))).)).)))))....)))))).....)))..((.(((((((...........))))))).))....... ( -48.50) >Bsubt.0 3172985 120 + 4214630/680-800 ACUACCCUGGCUGUAUUGACCUUCUAACCCCCCGCCCUUAUCGGGCGGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUU ....(((.((((((.((((((((((....((((((((.....))))))))))).)).)))))....)))))).....)))..((.(((((((...........))))))).))....... ( -53.40) >Banth.0 5077122 120 - 5227293/680-800 ACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUU (((..((((((....................((((((.....))))))(....)...))))))(((((.....(((((...)))))((((((...........)))))))))))..))). ( -50.10) >Bcere.0 5074106 120 - 5224283/680-800 ACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUU (((..((((((....................((((((.....))))))(....)...))))))(((((.....(((((...)))))((((((...........)))))))))))..))). ( -50.10) >consensus ACUACCCUGACUGUAUUGACAUUCUAACCUCCAGCCCUUAUCGGGCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAAGUAACGGAGGCGCCCAAAGGUU (((..((((((...((((....(((...((.((((((.....)))))))))))))))))))))(((((.....(((((...)))))((((((...........)))))))))))..))). (-48.79 = -44.97 + -3.83)

| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.44 |
| Mean single sequence MFE | -38.92 |
| Consensus MFE | -36.81 |
| Energy contribution | -33.35 |
| Covariance contribution | -3.46 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/720-840 CCCAGUCAAACUGCCCACCUGACACUGUCCCUGAACCGGAUCACGGUUCGAGGUUAGAAUGUCAAUACCGUCAGGGUAGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGCUUC ...((((..(((((((((.((((((((.((.(((((((.....))))))).)).)))..))))).....))..)))))))........(((((..............)))))..)))).. ( -36.94) >Bclau.0 2858068 120 - 4303871/720-840 CCCAGUCAAACUGCCCACCUGACAAUGUCCCUGGCCCGGAUCACGGGUCGAGGUUAGAAUGCCAGCACCAUCAGGGUAGUAUCCCACCGACGCCUCCACCGAAGCUAGCGCUCCGGUUUC .........(((((((...(((...(((...(((((((.....))))))).(((......))).)))...)))))))))).................((((.(((....))).))))... ( -34.90) >Blich.0 3118231 120 + 4222334/720-840 CCCAGUCAAACUGCCCACCUGACACUGUCUCCCUGCCCGAUAAGGGCAGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCACCGAAGCUGGCGCUCCGGUUUC .........(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).))))))).....((((..((..(((.......))).))..))))... ( -40.40) >Bsubt.0 3172985 120 + 4214630/720-840 CCCAGUCAAACUGCCCACCUGACACUGUCUCCCCGCCCGAUAAGGGCGGGGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCACCGAAGCUGGCGCUCCGGUUUC .........(((((((........(((.((((((((((.....)))))))))).)))..(((.......))).))))))).....((((..((..(((.......))).))..))))... ( -48.70) >Banth.0 5077122 120 - 5227293/720-840 CCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUC .........(((((((...((((.(((.((..((((((.....))))))..)).))).((....))...))))))))))).................((((.(((....))).))))... ( -36.30) >Bcere.0 5074106 120 - 5224283/720-840 CCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUC .........(((((((...((((.(((.((..((((((.....))))))..)).))).((....))...))))))))))).................((((.(((....))).))))... ( -36.30) >consensus CCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGCCCGAUAAGGGCCGGAGGUUAGAAUGUCAAUACAGUCAGGGUAGUAUCCCACCAACGCCUCCACCGAAGCUAGCGCUCCGGUUUC .........((((((((((((((...)))...((((((.....)))))).))))..(.....)..........))))))).................((((.(((....))).))))... (-36.81 = -33.35 + -3.46)

| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.44 |
| Mean single sequence MFE | -48.45 |
| Consensus MFE | -47.15 |
| Energy contribution | -42.47 |
| Covariance contribution | -4.69 |
| Combinations/Pair | 1.49 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/720-840 GAAGCCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACUACCCUGACGGUAUUGACAUUCUAACCUCGAACCGUGAUCCGGUUCAGGGACAGUGUCAGGUGGGCAGUUUGACUGGG ...((((.((((((.((....))...))))))(((((....)))))....)))).((((((((....(((.((((((.....)))))))))...)))))))).....(((.....))).. ( -46.80) >Bclau.0 2858068 120 - 4303871/720-840 GAAACCGGAGCGCUAGCUUCGGUGGAGGCGUCGGUGGGAUACUACCCUGAUGGUGCUGGCAUUCUAACCUCGACCCGUGAUCCGGGCCAGGGACAUUGUCAGGUGGGCAGUUUGACUGGG ...((((((((....)))))))).((((((((((.((.......))))))))(((....))).....))))..((((.....))))((((..(.((((((.....)))))).)..)))). ( -45.90) >Blich.0 3118231 120 + 4222334/720-840 GAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCUGCCCUUAUCGGGCAGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGG ...((((((((....))))))))(((((((.(((((((......)))..))))...)).)))))...((((((((((..(((.((((.(....)..)))).))))))))))......))) ( -49.80) >Bsubt.0 3172985 120 + 4214630/720-840 GAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCCCCGCCCUUAUCGGGCGGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGG ...((((((((....))))))))(((((((.(((((((......)))..))))...)).)))))...((((((((((.....)))))))).......((((..(.....)..)))).)). ( -52.40) >Banth.0 5077122 120 - 5227293/720-840 GAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGG ....((((((((((.((....))...))))))(((((....)))))(.((((((.((((....((......((((((.....))))))(....)))..))))....)))))).).)))). ( -47.90) >Bcere.0 5074106 120 - 5224283/720-840 GAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGG ....((((((((((.((....))...))))))(((((....)))))(.((((((.((((....((......((((((.....))))))(....)))..))))....)))))).).)))). ( -47.90) >consensus GAAACCGGAGCGCUAGCUUCGGUGGAGGCGCCGGUGGGAUACUACCCUGACUGUAUUGACAUUCUAACCUCCAGCCCUUAUCGGGCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGG ...((((((((....)))))))).......(((((.((((.(((((.((((...((((....(((...((.((((((.....))))))))))))))))))))))))...)))).))))). (-47.15 = -42.47 + -4.69)


| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.50 |
| Mean single sequence MFE | -50.37 |
| Consensus MFE | -52.01 |
| Energy contribution | -48.68 |
| Covariance contribution | -3.32 |
| Combinations/Pair | 1.47 |
| Mean z-score | -3.15 |
| Structure conservation index | 1.03 |
| SVM decision value | 4.00 |
| SVM RNA-class probability | 0.999748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/760-880 GGAUGUUGGUACCGUUUGUACAGGAUAGGUAGGAGCCUUGGAAGCCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACUACCCUGACGGUAUUGACAUUCUAACCUCGAACCGUGA ((((((..(((((((.....(((........((.(((((....((((((((....)))))))).))))).))(((((....)))))))))))))))..))))))................ ( -50.30) >Bclau.0 2858068 120 - 4303871/760-880 GAGUGUUGGUAUCAUCUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCUAGCUUCGGUGGAGGCGUCGGUGGGAUACUACCCUGAUGGUGCUGGCAUUCUAACCUCGACCCGUGA ((((((..((((((((...............(..(((((....((((((((....)))))))).)))))..)(((((....)))))..))))))))..))))))................ ( -51.30) >Blich.0 3118231 120 + 4222334/760-880 GAAUGUUGGUACAGCUUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCUGCCCUUA (((.((..((((((((.(..............(((((((....((((((((....)))))))).))))).))(((((....)))))).))))))))..)).)))................ ( -46.50) >Bsubt.0 3172985 120 + 4214630/760-880 GAAUGUUGGUACAGCUUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCCCCGCCCUUA (((.((..((((((((.(..............(((((((....((((((((....)))))))).))))).))(((((....)))))).))))))))..)).)))................ ( -46.50) >Banth.0 5077122 120 - 5227293/760-880 GAAUUUUGGUACAGUUUGUACAGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUA ((((((..(((((((.....((((.....(((..(((((....((((((((....)))))))).))))).)))((((....)))))))))))))))..))))))..(((....))).... ( -53.80) >Bcere.0 5074106 120 - 5224283/760-880 GAAUUUUGGUACAGUUUGUACAGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUA ((((((..(((((((.....((((.....(((..(((((....((((((((....)))))))).))))).)))((((....)))))))))))))))..))))))..(((....))).... ( -53.80) >consensus GAAUGUUGGUACAGUUUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCCGGUGGGAUACUACCCUGACUGUAUUGACAUUCUAACCUCCAGCCCUUA ((((((..((((((((...............((.(((((....((((((((....)))))))).))))).))(((((....)))))..))))))))..))))))................ (-52.01 = -48.68 + -3.32)

| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -25.99 |
| Energy contribution | -25.93 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/840-960 CAAGGCUCCUACCUAUCCUGUACAAACGGUACCAACAUCCAAUAUCAAGCUACAGUAAAGCUCCAUGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUU .((((((((..........((((.....))))...............((((.......))))....))))))))...(((((((.(((((...)))))......))))).))........ ( -25.90) >Bclau.0 2858068 120 - 4303871/840-960 CAAGGCUCCUACCUAUCCUGUACAGAUGAUACCAACACUCACUAUCAAGCUACAGUAAAGCUCCAUGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUU ..(((......)))..(((((..(((((.............((((..((((.......))))..))))(((...(((((......)))))..)))..)))))..)))))........... ( -27.60) >Blich.0 3118231 120 + 4222334/840-960 CAAGGCUCCUACCUAUCCUGUACAAGCUGUACCAACAUUCAAUAUCAGGCUGCAGUAAAGCUCCACGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUU ...((((..(((.......)))..))))(((((............((((..((.(.(((((((....))).))))).))))))..(((((...))))).........)))))........ ( -28.50) >Bsubt.0 3172985 120 + 4214630/840-960 CAAGGCUCCUACCUAUCCUGUACAAGCUGUACCAACAUUCAAUAUCAGGCUGCAGUAAAGCUCCACGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUU ...((((..(((.......)))..))))(((((............((((..((.(.(((((((....))).))))).))))))..(((((...))))).........)))))........ ( -28.50) >Banth.0 5077122 120 - 5227293/840-960 AAAGGCUCCCGCCUAUCCUGUACAAACUGUACCAAAAUUCAAUAUCAGGCUACAGUAAAGCUCCACGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUU ..((((....))))..(((((....((((((((..............)).))))))........((((((........)))))).(((((...)))))......)))))........... ( -29.44) >Bcere.0 5074106 120 - 5224283/840-960 AAAGGCUCCCGCCUAUCCUGUACAAACUGUACCAAAAUUCAAUAUCAGGCUACAGUAAAGCUCCACGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUU ..((((....))))..(((((....((((((((..............)).))))))........((((((........)))))).(((((...)))))......)))))........... ( -29.44) >consensus CAAGGCUCCUACCUAUCCUGUACAAACUGUACCAACAUUCAAUAUCAGGCUACAGUAAAGCUCCACGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUU .((((((((..........((((.....))))...............((((.......))))....))))))))...(((((((.(((((...)))))......))))).))........ (-25.99 = -25.93 + -0.05)

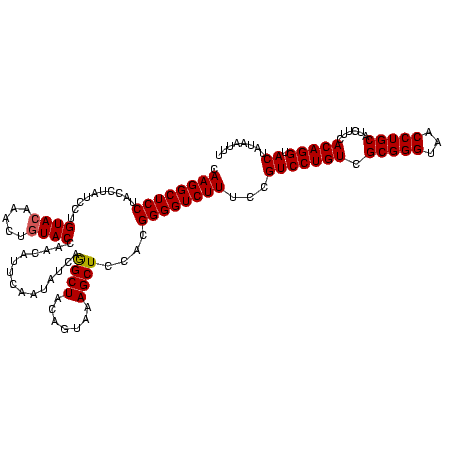
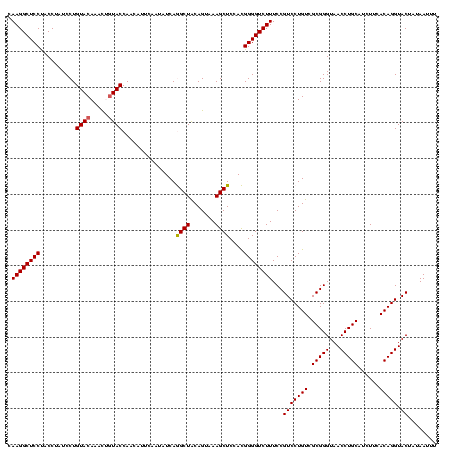
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -36.95 |
| Energy contribution | -36.23 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.15 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/920-1040 GCGAAAUUCCUUGUCGGGUAAGUUCCGACCCGCACGAAUGGUGUAACGACUUGGAUACUGUCUCAACGAGAGACCCGGUGAAAUUAUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAG (((....(((..(((((((........))))((((.....))))...)))..)))(((((((((.....)))))..))))........((((((((......))))).))).)))..... ( -37.20) >Bclau.0 2858068 120 - 4303871/920-1040 GCGAAAUUCCUUGUCGGGUAAGUUCCGACCCGCACGAAUGGUGCAACGACUUGGAUACUGUCUCAACGAGAGACCCGGUGAAAUUAUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAG (((....(((..(((((((........))))((((.....))))...)))..)))(((((((((.....)))))..))))........((((((((......))))).))).)))..... ( -39.60) >Blich.0 3118231 120 + 4222334/920-1040 GCGAAAUUCCUUGUCGGGUAAGUUCCGACCCGCACGAAAGGCGCAACGAUCUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUUAUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAG ((((........((((((.....)))))).(((.(....))))......)).((((((((((((.....)))))..))))..........((((((......))))))..)))))..... ( -37.40) >Bsubt.0 3172985 120 + 4214630/920-1040 GCGAAAUUCCUUGUCGGGUAAGUUCCGACCCGCACGAAAGGCGCAACGAUCUGGGCGCUGUCUCAACGAGAGACUCGGUGAAAUUAUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAG (((.........((((((.....)))))).(((.(((..(((((..........)))))(((((.....)))))))))))........((((((((......))))).))).)))..... ( -38.50) >Banth.0 5077122 120 - 5227293/920-1040 GCGAAAUUCCUUGUCGGGUAAGUUCCGACCCGCACGAAAGGUGUAACGAUUUGGGCACUGUCUCAACCAGAGACUCGGUGAAAUUAUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAG (((....(((..(((((((........))))((((.....))))...)))..)))(((((((((.....)))))..))))........((((((((......))))).))).)))..... ( -36.50) >Bcere.0 5074106 120 - 5224283/920-1040 GCGAAAUUCCUUGUCGGGUAAGUUCCGACCCGCACGAAAGGUGUAACGAUUUGGGCACUGUCUCAACCAGAGACUCGGUGAAAUUAUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAG (((....(((..(((((((........))))((((.....))))...)))..)))(((((((((.....)))))..))))........((((((((......))))).))).)))..... ( -36.50) >consensus GCGAAAUUCCUUGUCGGGUAAGUUCCGACCCGCACGAAAGGUGCAACGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUUAUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAG (((....(((..(((((((........))))((((.....))))...)))..)))(((((((((.....)))))..))))........((((((((......))))).))).)))..... (-36.95 = -36.23 + -0.72)
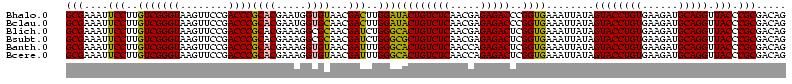


| Location | 3,178,984 – 3,179,103 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.58 |
| Mean single sequence MFE | -42.98 |
| Consensus MFE | -40.59 |
| Energy contribution | -38.23 |
| Covariance contribution | -2.35 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 119 - 4202352/1040-1160 GUAUAGGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAGGAGGGGUUAUCCCUU-CGGGAGAAGCUCUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUA ..............(((((((...((((((..((((..((...((((((.((((..-..)).))))))))...))..))))))))))...)))((((((.......))))))...)))). ( -41.70) >Bclau.0 2858068 120 - 4303871/1040-1160 GUAUAGGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAGGAAGGGUCAUCCCUUACGGGAGAAGCUCUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUA ..............(((((((...((((((..((((..((...(((((..((((....))))...)))))...))..))))))))))...)))((((((.......))))))...)))). ( -41.80) >Blich.0 3118231 116 + 4222334/1040-1160 GUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCG----UAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUA .....((((((((((.(((((...((((((..((((..((...((((((.((.----...))..))))))...))..))))))))))...))))).)))........)))))))...... ( -44.90) >Bsubt.0 3172985 116 + 4214630/1040-1160 GUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCG----UAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUA .....((((((((((.(((((...((((((..((((..((...((((((.((.----...))..))))))...))..))))))))))...))))).)))........)))))))...... ( -44.90) >Banth.0 5077122 116 - 5227293/1040-1160 GUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCG----UAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUA ........(....)(((((((...((((((..((((..((...((((((.((.----...))..))))))...))..))))))))))...)))((((((.......))))))...)))). ( -42.30) >Bcere.0 5074106 116 - 5224283/1040-1160 GUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCG----UAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUA ........(....)(((((((...((((((..((((..((...((((((.((.----...))..))))))...))..))))))))))...)))((((((.......))))))...)))). ( -42.30) >consensus GUAUAGGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAGGAGGGGUUAGCG____UAAGCGAAGCUCUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUA ..............(((((((...((((((..((((..((...(((((..((........))...)))))...))..))))))))))...)))((((((.......))))))...)))). (-40.59 = -38.23 + -2.35)
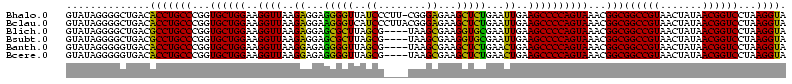
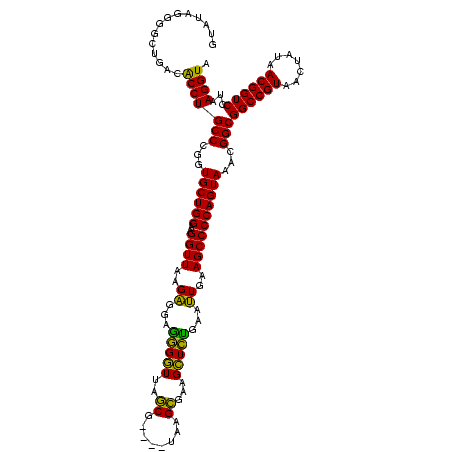
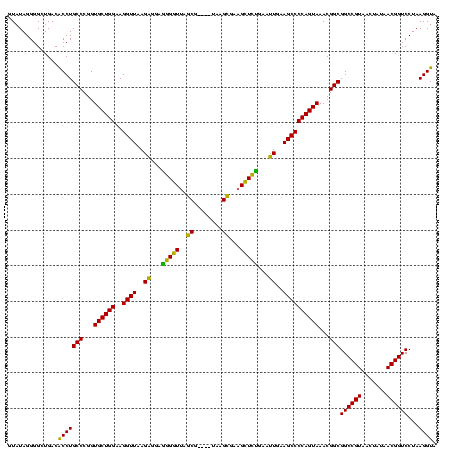
| Location | 3,178,984 – 3,179,103 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.84 |
| Mean single sequence MFE | -37.98 |
| Consensus MFE | -35.30 |
| Energy contribution | -32.50 |
| Covariance contribution | -2.80 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 119 - 4202352/1080-1200 UUUAGCAAAAACACAGGUCUCUGCGAAGCCGCAAGGCGAAGUAUAGGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAGGAGGGGUUAUCCCUU-CGGGAGAAGCUCUGAAUUGAAGC .............((((..(((.(...(((....)))..(((((..((((........)))).))))).............((((((....)))))-).).)))..).)))......... ( -38.50) >Bclau.0 2858068 120 - 4303871/1080-1200 UUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAGGAAGGGUCAUCCCUUACGGGAGAAGCUCUGAAUUGAAGC .............((((..(((.(...(((....)))...(((.(((((.((((.(((.(((...(((....)))...).)).))))))))))))))).).)))..).)))......... ( -38.80) >Blich.0 3118231 116 + 4222334/1080-1200 UUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCG----UAAGCGAAGGUGCGAAUUGAAGC (((((((...........((.(((...(((....)))...))).))((((........))))..)))))))..(((..((...((((((.((.----...))..))))))...))..))) ( -37.90) >Bsubt.0 3172985 116 + 4214630/1080-1200 UUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCG----UAAGCGAAGGUGCGAAUUGAAGC (((((((...........((.(((...(((....)))...))).))((((........))))..)))))))..(((..((...((((((.((.----...))..))))))...))..))) ( -37.90) >Banth.0 5077122 116 - 5227293/1080-1200 UUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCG----UAAGCGAAGCUCUGAACUGAAGC .............(((((((((....((((....((((..(..((((.(....).))))..)..))))....))))..))))))(((((.((.----...))..)))))....))).... ( -37.40) >Bcere.0 5074106 116 - 5224283/1080-1200 UUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCG----UAAGCGAAGCUCUGAACUGAAGC .............(((((((((....((((....((((..(..((((.(....).))))..)..))))....))))..))))))(((((.((.----...))..)))))....))).... ( -37.40) >consensus UUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAGGAGGGGUUAGCG____UAAGCGAAGCUCUGAAUUGAAGC ((((((.........(((((((((...(((....)))...))..)))))))...(((.....)))))))))..(((..((...(((((..((........))...)))))...))..))) (-35.30 = -32.50 + -2.80)

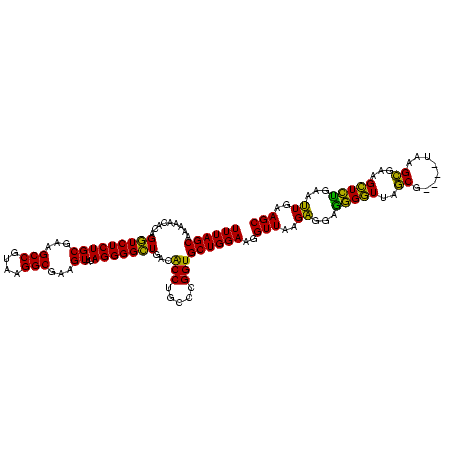
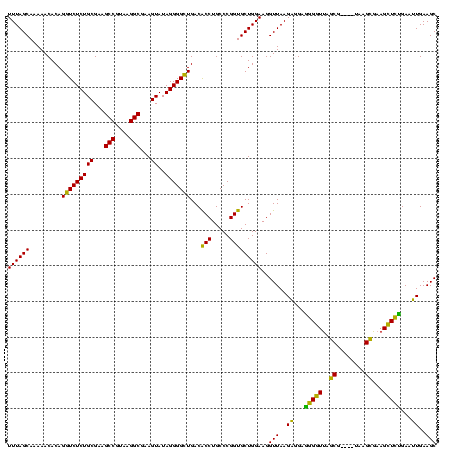
| Location | 3,178,984 – 3,179,103 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.53 |
| Mean single sequence MFE | -41.73 |
| Consensus MFE | -35.78 |
| Energy contribution | -36.53 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 119 - 4202352/1160-1280 ACCCCGUAACUUCGGGAGAAGGGGUGCUCUGAUAGGGUGCA-AGCCCGAGAGAGCCGCAGUGAAAAGAUCCAAGCGACUGUUUAGCAAAAACACAGGUCUCUGCGAAGCCGCAAGGCGAA (((((....(....).....)))))(((((....((((...-.))))...)))))(((((.((..........((.........))...........)).)))))..(((....)))... ( -41.70) >Bclau.0 2858068 120 - 4303871/1160-1280 ACCCCGUAACUUCGGGAGAAGGGGUGCUCUAGUAGGGUUAAUCGCCCGAGAGAGCCGCAGUGAAAGGAUCCAAGCGACUGUUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAA (((((....(....).....)))))(((((....((((.....))))...)))))(((((.((..(....)......((((...........)))).)).)))))..(((....)))... ( -42.10) >Blich.0 3118231 119 + 4222334/1160-1280 ACCCCGUAACUUCGGGAGAAGGGGUGCUCUGUUAGGGUGCA-AGCCCGAGAGAGCCGCAGUGAAUAGGCCCAGGCGACUGUUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAA (((((....(....).....)))))(((((.((.((((...-.)))))).)))))(((((.....(((((.((....))((((.....))))...))))))))))..(((....)))... ( -46.60) >Bsubt.0 3172985 119 + 4214630/1160-1280 ACCCCGUAACUUCGGGAGAAGGGGUGCUCUGUUAGGGUGCA-AGCCCGAGAGAGCCGCAGUGAAUAGGCCCAGGCGACUGUUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAA (((((....(....).....)))))(((((.((.((((...-.)))))).)))))(((((.....(((((.((....))((((.....))))...))))))))))..(((....)))... ( -46.60) >Banth.0 5077122 108 - 5227293/1160-1280 ACCCCGUAACUUCGGGAGAAGGGGUGCUUUCUUAACG------------GAAAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAA (((((....(....).....)))))(((((((....)------------))))))((((((((((((((....))..)))))))((.........))...)))))..(((....)))... ( -36.70) >Bcere.0 5074106 108 - 5224283/1160-1280 ACCCCGUAACUUCGGGAGAAGGGGUGCUUUCUUAACG------------GAAAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAA (((((....(....).....)))))(((((((....)------------))))))((((((((((((((....))..)))))))((.........))...)))))..(((....)))... ( -36.70) >consensus ACCCCGUAACUUCGGGAGAAGGGGUGCUCUGUUAGGGUGCA_AGCCCGAGAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAA (((((....(....).....)))))(((((....((((.....))))...)))))(((((.((....((....))..((((...........)))).)).)))))..(((....)))... (-35.78 = -36.53 + 0.75)
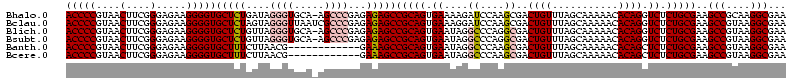
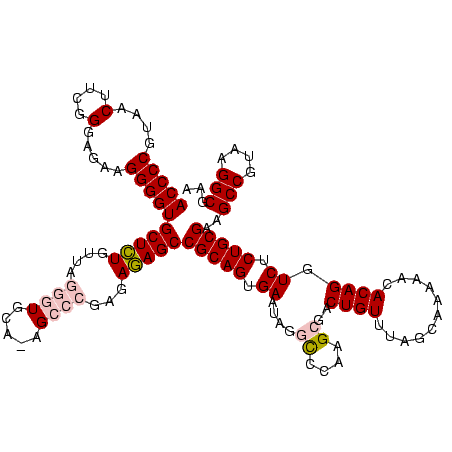

| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -30.83 |
| Energy contribution | -30.67 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/1280-1400 CAUUUUGCCGAGUUCCUUAACGAGAGUUCUCCCGAGCGUCUUAGAAUUCUCUUCUCGCCUACCUGUGUCGGUUUGCGGUACGGGCACCUCUCACCUCGCUAGAGGCUUUUCUUGGCAGUG ....((((((((.........(((((....((((.((.....((((.....))))(((..(((......)))..))))).))))...))))).((((....)))).....)))))))).. ( -39.10) >Bclau.0 2858068 120 - 4303871/1280-1400 CAUUUUGCCGAGUUCCUUAACGAGAGUUCUCCCGAGCGUCUUAGAAUUCUCUUCUCGCCUACCUGUGUCGGUUUGCGGUACGGGCACCUGUAUUCUCACUAGAGGCUUUUCUCGGCAGCG ....((((((((..((((...(((((((((...(((...))))))))))))....(((..(((......)))..)))((((((....))))))........)))).....)))))))).. ( -38.20) >Blich.0 3118231 120 + 4222334/1280-1400 CAUUUUGCCGAGUUCCUUAACGAGAGUUCUCUCGAUCACCUUAGGAUUCUCUCCUCGCCUACCUGUGUCGGUUUGCGGUACGGGCACCUCUCACCUCGCUAGAGGCUUUUCUUGGCAGUG ....((((((((.........(((((....((((...(((..((((.....)))).((..(((......)))..))))).))))...))))).((((....)))).....)))))))).. ( -37.20) >Bsubt.0 3172985 120 + 4214630/1280-1400 CAUUUUGCCGAGUUCCUUAACGAGAGUUCUCUCGAUCACCUUAGGAUUCUCUCCUCGCCUACCUGUGACGGUUUGCGGUACGGGCACCUCUCACCUCGCUAGAGGCUUUUCUUGGCAGUG ....((((((((.........(((((((((............))))))))).....((((....((((.(((..(.(((......))).)..)))))))...))))....)))))))).. ( -37.20) >Banth.0 5077122 120 - 5227293/1280-1400 CAUUUUGCCGAGUUCCUUAACCAGAGUUCUCUCGCACACCUUAGGAUUCUCUCCUCGCCUACCUGUGUCGGUUUGCGGUACAGGCACCUUUUAUCUCGCUAGAAGCUUUUCUUGGCAGCG ....((((((((..........((((((.(((.((.......((((.....))))......(((((..((.....))..))))).............)).))))))))).)))))))).. ( -30.50) >Bcere.0 5074106 120 - 5224283/1280-1400 CAUUUUGCCGAGUUCCUUAACCAGAGUUCUCUCGCACACCUUAGGAUUCUCUCCUCGCCUACCUGUGUCGGUUUGCGGUACAGGCACCUUUUAUCUCGCUAGAAGCUUUUCUUGGCAGCG ....((((((((..........((((((.(((.((.......((((.....))))......(((((..((.....))..))))).............)).))))))))).)))))))).. ( -30.50) >consensus CAUUUUGCCGAGUUCCUUAACGAGAGUUCUCUCGAACACCUUAGGAUUCUCUCCUCGCCUACCUGUGUCGGUUUGCGGUACGGGCACCUCUCACCUCGCUAGAGGCUUUUCUUGGCAGCG ....((((((((.........(((((((((............)))))))))..........(((((..((.....))..))))).........((((....)))).....)))))))).. (-30.83 = -30.67 + -0.17)
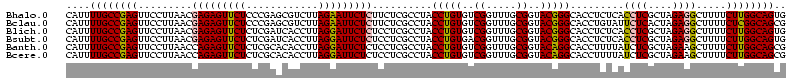
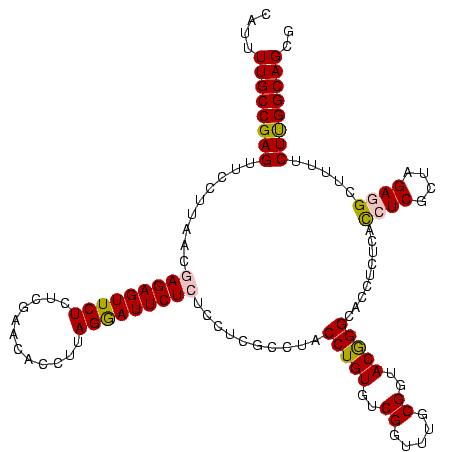
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.05 |
| Mean single sequence MFE | -39.65 |
| Consensus MFE | -36.51 |
| Energy contribution | -35.57 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/1600-1720 GCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUGGGCUUUCGACGGAGGGGAUUCUCACCCCUCUUUUCGCUACUCAUACCGGCAUUCU (((....((((((.(((((((...........))))))).))((((..((((...))))..))))))))..(((.(((((((.......)))))))..)))...........)))..... ( -41.20) >Bclau.0 2858068 120 - 4303871/1600-1720 GCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUGGGCUUUCGACGGAUAGGAUUCUCACCUAUCUUUUCGCUACUCAUACCGGCAUUCU (((....((((((.(((((((...........))))))).))((((..((((...))))..))))))))..(((.(((((((.......)))))))..)))...........)))..... ( -37.00) >Blich.0 3118231 118 + 4222334/1600-1720 GCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAACC-UUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUCUUU-CGCUACUCAUACCGGCAUUCU (((....((((((.(((((((...........))))))).)).).)))..(((.((....)-).)))....(((.(((((((.......))))))).)-))...........)))..... ( -40.30) >Bsubt.0 3172985 119 + 4214630/1600-1720 GCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAACCCUUAGGCAUUCGGUGGAGGG-AUUCUCACCCCUCUUUUCGCUACUCAUACCGGCAUUCU (((....((((((.(((((((...........))))))).)).).)))..(((.(((...))).)))....(((.((((((-........))))))..)))...........)))..... ( -37.00) >Banth.0 5077122 119 - 5227293/1600-1720 GCCUGUCGGCCUCGGCUUAGGUCCUGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUCUUU-CGCUACUCAUACCGGCAUUCU (((((((((((((.(((((((...........))))))).)).).)))((((...))))......))))..(((.(((((((.......))))))).)-))...........)))..... ( -41.20) >Bcere.0 5074106 119 - 5224283/1600-1720 GCCUGUCGGCCUCGGCUUAGGUCCUGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUCUUU-CGCUACUCAUACCGGCAUUCU (((((((((((((.(((((((...........))))))).)).).)))((((...))))......))))..(((.(((((((.......))))))).)-))...........)))..... ( -41.20) >consensus GCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUCUUU_CGCUACUCAUACCGGCAUUCU ((((...((((((.(((((((...........))))))).)).).)))((((...)))).....)))).......(((((((.......)))))))....(((.........)))..... (-36.51 = -35.57 + -0.94)

| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.84 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -27.26 |
| Energy contribution | -25.73 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/1680-1800 CACCGUUGGUGCGAGUGGUAGGGGAGCGUUCUACGUGCAGCGAAGUCAGACCGAGAGGACUGGUGGAGCGCGUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAAGAGGGGUGAGAAUCC ((((.(((.(((.(.(((((.....((.((((((((((..((...((((.((....)).))))))..)))))))))))).....))))).)....))).))).......))))....... ( -38.50) >Bclau.0 2858068 119 - 4303871/1680-1800 UCCCGUAAGGGAUUG-GGUAGGGGAGCGUUCCAAGGACAGCGAAGCUAGAUCGUGAGGACUGGUGGAGUGCUUGGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAAGAUAGGUGAGAAUCC ((((....))))...-((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))................................ ( -34.40) >Blich.0 3118231 116 + 4222334/1680-1800 UCU---UGCGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCGAGACCGGAAGGACUCGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCG-AAAGAGGGGUGAGAAUCC (((---..(....(.(((((.....((.(((((((.(((.(...(((((.((....)).)))))).))).))))))))).....))))).)........(.-...)....)..))).... ( -39.50) >Bsubt.0 3172985 115 + 4214630/1680-1800 UCU---U-CGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCCAGACCGGAAGGACUGGUGGACGGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAAGAGGGGUGAGAAU-C (((---(-(.......((((.....((.(((((..((((((...(((((.((....)).)))))..))))))))))))).....))))(.((....)).).....)))))........-. ( -34.70) >Banth.0 5077122 119 - 5227293/1680-1800 UACCAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCG-AAAGACGGGUGAGAAUCC ((((.......(((.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))))...)))...((.-...).).))))....... ( -30.30) >Bcere.0 5074106 119 - 5224283/1680-1800 UACCAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCG-AAAGACGGGUGAGAAUCC ((((.......(((.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))))...)))...((.-...).).))))....... ( -30.30) >consensus UACC__UGGGAACAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGCCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCG_AAAGAGGGGUGAGAAUCC (((.....))).....((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))....................((((....)))) (-27.26 = -25.73 + -1.52)

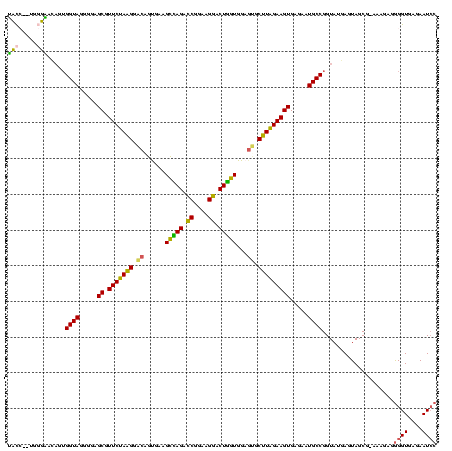
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.58 |
| Mean single sequence MFE | -30.14 |
| Consensus MFE | -22.50 |
| Energy contribution | -21.34 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/1720-1840 CACUUCUACGCGCUCCACCAGUCCUCUCGGUCUGACUUCGCUGCACGUAGAACGCUCCCCUACCACUCGCACCAACGGUGCGAAUCCAUAGCUUCGGUGAUACGUUUAGCCCCGGUACAU ...(((((((.((.....(((.((....)).)))........)).)))))))........((((..(((((((...))))))).......(((.((......))...)))...))))... ( -34.32) >Bclau.0 2858068 117 - 4303871/1720-1840 CACUUCCAAGCACUCCACCAGUCCUCACGAUCUAGCUUCGCUGUCCUUGGAACGCUCCCCUACC-CAAUCCCUUACGGGAUUG--CCAUAGCUUCGGUGAUACGUUUAGCCCCGGUACAU ...(((((((.((.......(((.....)))..(((...))))).)))))))........((((-(((((((....)))))))--.....(((.((......))...)))...))))... ( -25.60) >Blich.0 3118231 117 + 4222334/1720-1840 CACUUCUAAGCGCUCCACGAGUCCUUCCGGUCUCGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCGCA---AGAACAGUCCGCAGCUUCGGUGAUACGUUUAGCCCCGGUACAU ...(((((((.(((...((((.((....)).))))......))).)))))))............(((((((...---.)))))))(((..(((.((......))...)))..)))..... ( -36.60) >Bsubt.0 3172985 116 + 4214630/1720-1840 CACUUCUAAGCCGUCCACCAGUCCUUCCGGUCUGGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCG-A---AGAACAGUCCGCAGCUUCGGUGAUACGUUUAGCCCCGGUACAU ...((((((((.((...((((.((....)).))))....)).))..))))))............(((((((.-.---.)))))))(((..(((.((......))...)))..)))..... ( -27.90) >Banth.0 5077122 120 - 5227293/1720-1840 CACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUUGGUAUCAAUCCGCAGCUUCGGUGGUGUAUUUAGCCCCGGUACAU ...(((((((.((.....(((.((....)).)))........)).)))))))........((((..(((((((...)))))))............((.(((.......)))))))))... ( -28.22) >Bcere.0 5074106 120 - 5224283/1720-1840 CACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUUGGUAUCAAUCCGCAGCUUCGGUGGUGUAUUUAGCCCCGGUACAU ...(((((((.((.....(((.((....)).)))........)).)))))))........((((..(((((((...)))))))............((.(((.......)))))))))... ( -28.22) >consensus CACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGCCCUUAGAACGCUCCCCUACCACUGUUACCA__GGGAUCAAUCCGCAGCUUCGGUGAUACGUUUAGCCCCGGUACAU ...(((((((.((.....(((.((....)).)))........)).)))))))........((((..((((((.....)))))).......(((.((......))...)))...))))... (-22.50 = -21.34 + -1.16)
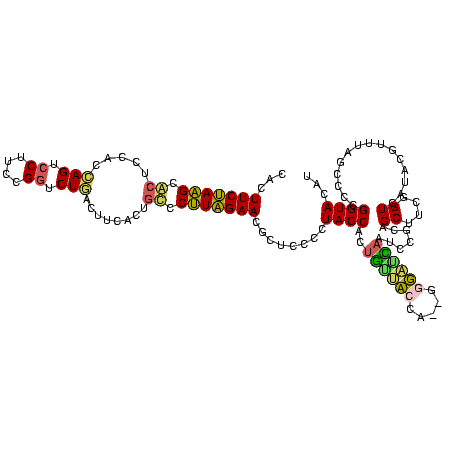
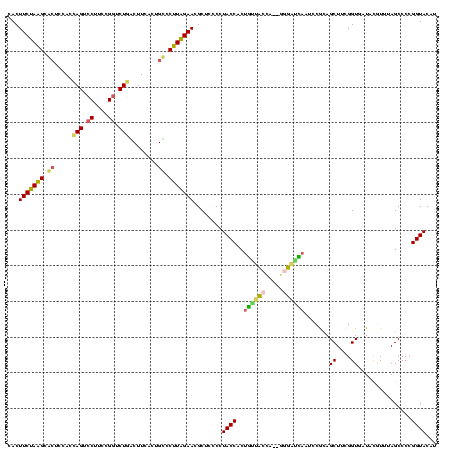
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.58 |
| Mean single sequence MFE | -40.96 |
| Consensus MFE | -38.11 |
| Energy contribution | -35.35 |
| Covariance contribution | -2.76 |
| Combinations/Pair | 1.51 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/1720-1840 AUGUACCGGGGCUAAACGUAUCACCGAAGCUAUGGAUUCGCACCGUUGGUGCGAGUGGUAGGGGAGCGUUCUACGUGCAGCGAAGUCAGACCGAGAGGACUGGUGGAGCGCGUAGAAGUG ..........(((...(.((((.(((......)))(((((((((...))))))))))))).)..))).((((((((((..((...((((.((....)).))))))..))))))))))... ( -44.20) >Bclau.0 2858068 117 - 4303871/1720-1840 AUGUACCGGGGCUAAACGUAUCACCGAAGCUAUGG--CAAUCCCGUAAGGGAUUG-GGUAGGGGAGCGUUCCAAGGACAGCGAAGCUAGAUCGUGAGGACUGGUGGAGUGCUUGGAAGUG ..........(((...(.((((.(((......)))--(((((((....)))))))-)))).)..))).(((((((.((......(((((.((....)).)))))...)).)))))))... ( -41.50) >Blich.0 3118231 117 + 4222334/1720-1840 AUGUACCGGGGCUAAACGUAUCACCGAAGCUGCGGACUGUUCU---UGCGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCGAGACCGGAAGGACUCGUGGAGCGCUUAGAAGUG .....(((.((((...((......)).)))).)))(((((((.---...))))))).........((.(((((((.(((.(...(((((.((....)).)))))).))).))))))))). ( -44.70) >Bsubt.0 3172985 116 + 4214630/1720-1840 AUGUACCGGGGCUAAACGUAUCACCGAAGCUGCGGACUGUUCU---U-CGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCCAGACCGGAAGGACUGGUGGACGGCUUAGAAGUG ..((.(((.((((...((......)).)))).))))).(((((---(-(..((....)).))))))).(((((..((((((...(((((.((....)).)))))..)))))))))))... ( -43.30) >Banth.0 5077122 120 - 5227293/1720-1840 AUGUACCGGGGCUAAAUACACCACCGAAGCUGCGGAUUGAUACCAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUG .....(((.((((..............)))).)))(((((((((...))))))))).........((.(((((((.((......(((((.((....)).)))))...)).))))))))). ( -36.04) >Bcere.0 5074106 120 - 5224283/1720-1840 AUGUACCGGGGCUAAAUACACCACCGAAGCUGCGGAUUGAUACCAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUG .....(((.((((..............)))).)))(((((((((...))))))))).........((.(((((((.((......(((((.((....)).)))))...)).))))))))). ( -36.04) >consensus AUGUACCGGGGCUAAACGUAUCACCGAAGCUGCGGAUUGAUACC__UGGGAACAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGCCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUG ..........(((...(.((((.(((......)))((((((((.....)))))))))))).)..))).(((((((.((......(((((.((....)).)))))...)).)))))))... (-38.11 = -35.35 + -2.76)

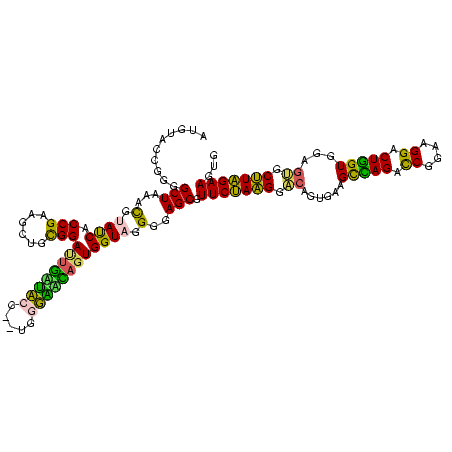
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -25.33 |
| Consensus MFE | -24.60 |
| Energy contribution | -23.93 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/1840-1960 UUUCGGCGCAGAGUCACUCGACCAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUAAGCCAACAUCCUGGUUGUCUGGGCAACUCCACAUCCUUUUCCACUUAACGUAUACUU .....((.((((......(((((((.((((......))..........((.(((((.......))))).))))))))))))))).))................................. ( -27.90) >Bclau.0 2858068 120 - 4303871/1840-1960 UUUCGGCGCAGAGUCACUCGACCAGUGAGCUAUUACGCACUCUUUCAAUGAUGGCUGCUUCUAAGCCAACAUCCUGGUUGUCUGGGCAACUCCACAUCCUUUUCCACUUAACGUAUACUU .....((.((((......(((((((.((((......))..........((.(((((.......))))).))))))))))))))).))................................. ( -27.90) >Blich.0 3118231 120 + 4222334/1840-1960 UUUCGGCGCAGAGUCACUCGACCAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUAAGCCAACAUCCUGGUUGUCUAAGCAACUCCACAUCCUUUUCCACUUAACGUAUACUU .....(((.(((((((((.....)))))((......)).)))).......((((.(((((...(((((......)))))....)))))...))))................)))...... ( -24.90) >Bsubt.0 3172985 120 + 4214630/1840-1960 UUUCGGCGCAGAGUCACUCGACCAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUAAGCCAACAUCCUGGUUGUCUAAGCAACUCCACAUCCUUUUCCACUUAACGUAUACUU .....(((.(((((((((.....)))))((......)).)))).......((((.(((((...(((((......)))))....)))))...))))................)))...... ( -24.90) >Banth.0 5077122 120 - 5227293/1840-1960 UUUCGGCGCAGAGUCACUCGACUAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUAAGCCAACAUCCUAGUUGUCUAAGCAACUCCACAUCCUUUUCCACUUAAUACACACUU .....(((...(((((((.....))))).))....))).....((((.((((((((.......))))).......((((((....))))))............))).))))......... ( -23.20) >Bcere.0 5074106 120 - 5224283/1840-1960 UUUCGGCGCAGAGUCACUCGACUAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUAAGCCAACAUCCUAGUUGUCUAAGCAACUCCACAUCCUUUUCCACUUAAUACACACUU .....(((...(((((((.....))))).))....))).....((((.((((((((.......))))).......((((((....))))))............))).))))......... ( -23.20) >consensus UUUCGGCGCAGAGUCACUCGACCAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUAAGCCAACAUCCUGGUUGUCUAAGCAACUCCACAUCCUUUUCCACUUAACGUAUACUU ....((....((((..(((((((((.((((......))..........((.(((((.......))))).)))))))))))....))..)))).....))..................... (-24.60 = -23.93 + -0.67)

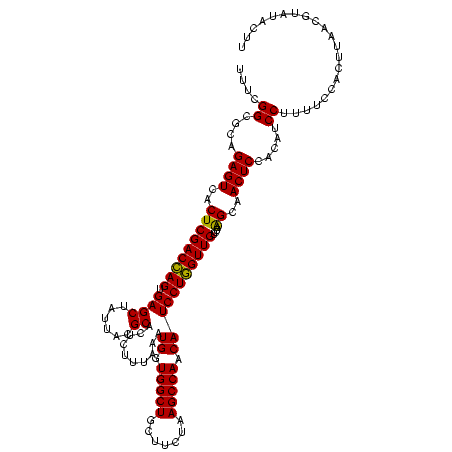
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -36.68 |
| Energy contribution | -36.35 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.62 |
| SVM RNA-class probability | 0.999930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/1840-1960 AAGUAUACGUUAAGUGGAAAAGGAUGUGGAGUUGCCCAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAA .....(((.....))).....((.((..((((..(...((...((((((((.(((((.......))))).))))....((((........)))).))))))..)..))))..)))).... ( -35.50) >Bclau.0 2858068 120 - 4303871/1840-1960 AAGUAUACGUUAAGUGGAAAAGGAUGUGGAGUUGCCCAGACAACCAGGAUGUUGGCUUAGAAGCAGCCAUCAUUGAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAA .....(((.....))).....((.((..((((..(...((...((((((((.(((((.......))))).))))....((((........)))).))))))..)..))))..)))).... ( -36.20) >Blich.0 3118231 120 + 4222334/1840-1960 AAGUAUACGUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAA .....(((.....))).....((.((..((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..))))..)))).... ( -38.20) >Bsubt.0 3172985 120 + 4214630/1840-1960 AAGUAUACGUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAA .....(((.....))).....((.((..((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..))))..)))).... ( -38.20) >Banth.0 5077122 120 - 5227293/1840-1960 AAGUGUGUAUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACUAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAA .....................((.((..((((..(((.(((.....(((((.(((((.......))))).)))))....((((.((....)).)))).))))))..))))..)))).... ( -37.10) >Bcere.0 5074106 120 - 5224283/1840-1960 AAGUGUGUAUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACUAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAA .....................((.((..((((..(((.(((.....(((((.(((((.......))))).)))))....((((.((....)).)))).))))))..))))..)))).... ( -37.10) >consensus AAGUAUACGUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAA .....................((.((..((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..))))..)))).... (-36.68 = -36.35 + -0.33)
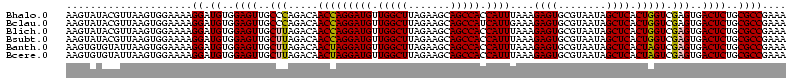
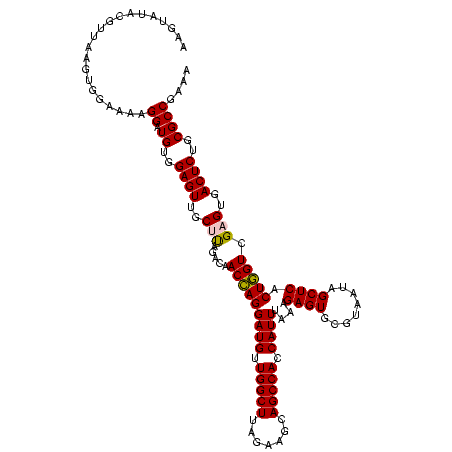
| Location | 3,178,984 – 3,179,103 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.66 |
| Mean single sequence MFE | -41.45 |
| Consensus MFE | -39.24 |
| Energy contribution | -37.85 |
| Covariance contribution | -1.39 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 119 - 4202352/2120-2240 GCCCGAACCCACGCAUGUUGAAAAAUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACUCGGAAAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUCGAGGG-AA .(((...(((.(((((........)))))))).((((((.......((((.(((..((...(((....))).....))...))).))))....(((((....))))))))))).)))-.. ( -44.40) >Bclau.0 2858068 119 - 4303871/2120-2240 GCCCGAACCCACGCAAGUUGAAAAUUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACUCGGAAAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUCGAGUG-AU ((((...(((.(((((........)))))))).((((((.......((((.(((..((...(((....))).....))...))).))))......))))))))))............-.. ( -38.92) >Blich.0 3118231 119 + 4222334/2120-2240 GCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAAG-GUAA (((....(((.(((((........)))))))).((((((.((((((((..(((........)))..))))((((........))))))))...(((((....))))))))))).)-)).. ( -42.60) >Bsubt.0 3172985 119 + 4214630/2120-2240 GCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAAG-GUAA (((....(((.(((((........)))))))).((((((.((((((((..(((........)))..))))((((........))))))))...(((((....))))))))))).)-)).. ( -42.60) >Banth.0 5077122 120 - 5227293/2120-2240 GCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUUAAGUGUAA ((((...(((.(((((........)))))))).((((((.((((....((((...))))....(((((.((......)))))))..)))).....))))))))))............... ( -40.10) >Bcere.0 5074106 120 - 5224283/2120-2240 GCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUUAAGUGUAA ((((...(((.(((((........)))))))).((((((.((((....((((...))))....(((((.((......)))))))..)))).....))))))))))............... ( -40.10) >consensus GCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUCAAGUGUAA ((((...(((.(((((........)))))))).((((((.((((....(((.....)))....(((((.((......)))))))..)))).....))))))))))............... (-39.24 = -37.85 + -1.39)

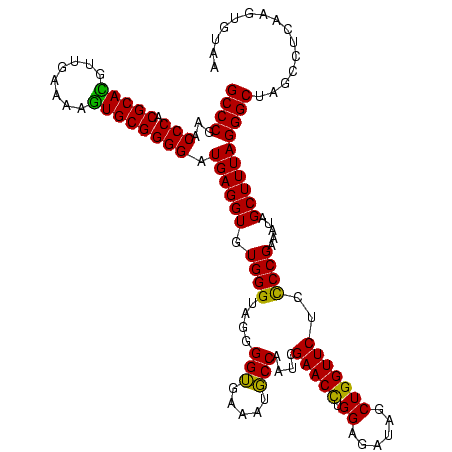
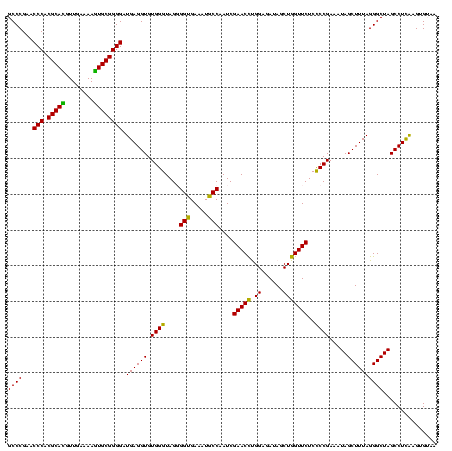
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -33.38 |
| Energy contribution | -31.88 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/2160-2280 CAGCUAUUUCCGAGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCAUUUUUCAACAUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGU ..((((....((....))..))))...........(((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).))))) ( -31.70) >Bclau.0 2858068 120 - 4303871/2160-2280 CAGCUAUUUCCGAGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCAAUUUUCAACUUGCGUGGGUUCGGGCCUCCAGUCGGUGUUACCCGACCUUCACCCUGGACAUGGGU ..((((....((....))..))))...........(((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).))))) ( -35.20) >Blich.0 3118231 120 + 4222334/2160-2280 CAGCUAUCUCCAGGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGU ...((((.((((((......(((.......(((..((((((...........((((........))))))))))..)))...)))(((((......)))))......)))))).)))).. ( -34.90) >Bsubt.0 3172985 120 + 4214630/2160-2280 CAGCUAUCUCCAGGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGU ...((((.((((((......(((.......(((..((((((...........((((........))))))))))..)))...)))(((((......)))))......)))))).)))).. ( -34.90) >Banth.0 5077122 120 - 5227293/2160-2280 CAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGU ...((((.((((((......((((.....((((..((((((...........((((........)))))))))).))))..))))(((((......)))))......)))))).)))).. ( -36.60) >Bcere.0 5074106 120 - 5224283/2160-2280 CAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGU ...((((.((((((......((((.....((((..((((((...........((((........)))))))))).))))..))))(((((......)))))......)))))).)))).. ( -36.60) >consensus CAGCUAUCUCCAGGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGU .........((((......))))............(((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).))))) (-33.38 = -31.88 + -1.50)
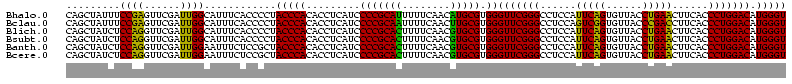
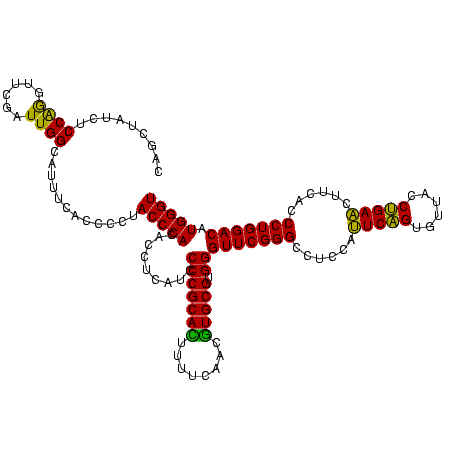
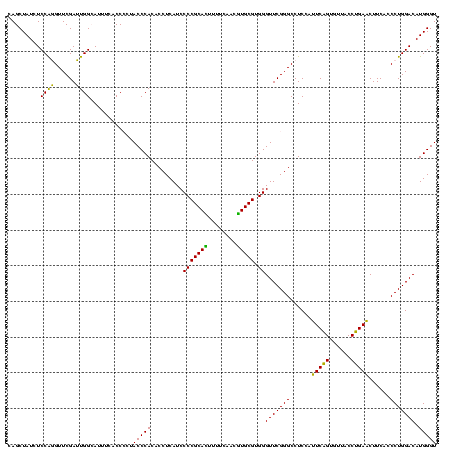
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -45.17 |
| Consensus MFE | -44.33 |
| Energy contribution | -42.83 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.45 |
| SVM RNA-class probability | 0.999987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/2160-2280 ACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCAUGUUGAAAAAUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACUCGGAAAUAGCUG ((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..))))))...(((.....)))..(((....)))......... ( -42.30) >Bclau.0 2858068 120 - 4303871/2160-2280 ACCCAUGUCCAGGGUGAAGGUCGGGUAACACCGACUGGAGGCCCGAACCCACGCAAGUUGAAAAUUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACUCGGAAAUAGCUG ((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..))))))...(((.....)))..(((....)))......... ( -45.30) >Blich.0 3118231 120 + 4222334/2160-2280 ACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUG ((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..))))))(((..(((........)))..)))........... ( -46.00) >Bsubt.0 3172985 120 + 4214630/2160-2280 ACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUG ((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..))))))(((..(((........)))..)))........... ( -46.00) >Banth.0 5077122 120 - 5227293/2160-2280 ACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUG ((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..))))))(((.......(((((........)))))...))). ( -45.70) >Bcere.0 5074106 120 - 5224283/2160-2280 ACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUG ((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..))))))(((.......(((((........)))))...))). ( -45.70) >consensus ACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUG ((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..))))))...(((.....)))..................... (-44.33 = -42.83 + -1.50)

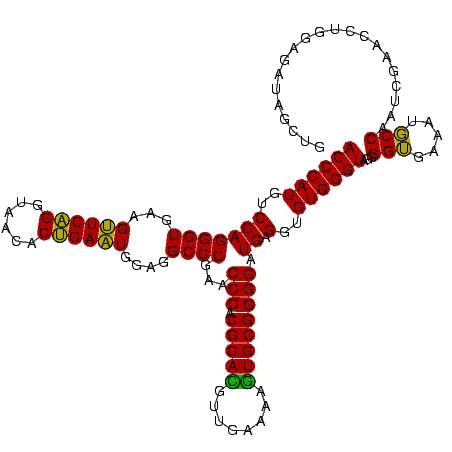
| Location | 3,178,984 – 3,179,103 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.52 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -34.21 |
| Energy contribution | -33.10 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 119 - 4202352/2200-2320 CACCUCAUCCCCGCAUUUUUCAACAUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACACGGUUUCGGGUCUACGACGGCGUACUAUUU-C ..((....((((((((........))))).)))...))((((((((((((......)))))........)))).....(((((((.(........))))))))...))).........-. ( -31.30) >Bclau.0 2858068 120 - 4303871/2200-2320 CACCUCAUCCCCGCAAUUUUCAACUUGCGUGGGUUCGGGCCUCCAGUCGGUGUUACCCGACCUUCACCCUGGACAUGGGUAGAUCACCCGGUUUCGGGUCUACGGCAUCGUACUAAAGGC ..((....((((((((........))))).)))...))((((..((((((((((((((((((........))...(((((.....)))))...))))))....))))))).)))..)))) ( -43.90) >Blich.0 3118231 119 + 4222334/2200-2320 CACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGUCUACGACCACGUACUCAUG-C ........((((((((........))))).)))((((((......(((((......)))))......))))))((((((((((((....)))).((((((...)))).))))))))))-. ( -36.50) >Bsubt.0 3172985 119 + 4214630/2200-2320 CACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGUCUACGACCACGUACUCAUG-C ........((((((((........))))).)))((((((......(((((......)))))......))))))((((((((((((....)))).((((((...)))).))))))))))-. ( -36.50) >Banth.0 5077122 118 - 5227293/2200-2320 CACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGUCUACGACCACGUACU-AAA-C ........................(((((((((((((((......(((((......)))))......)))))))....(((((...((((....)))))))))..)))))))).-...-. ( -36.00) >Bcere.0 5074106 118 - 5224283/2200-2320 CACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGUCUACGACCACGUACU-AAA-C ........................(((((((((((((((......(((((......)))))......)))))))....(((((...((((....)))))))))..)))))))).-...-. ( -36.00) >consensus CACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGUCUACGACCACGUACU_AAG_C ..(((...((((((((........))))).)))...)))......(((((......)))))....((((..(((.(((((.....))))))))..)))).((((....))))........ (-34.21 = -33.10 + -1.11)
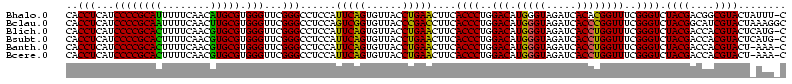
| Location | 3,178,984 – 3,179,103 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.52 |
| Mean single sequence MFE | -42.47 |
| Consensus MFE | -39.30 |
| Energy contribution | -39.08 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 119 - 4202352/2200-2320 G-AAAUAGUACGCCGUCGUAGACCCGAAACCGUGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCAUGUUGAAAAAUGCGGGGAUGAGGUG .-........((((...(((((((((....)).).).))))).(((.....((((...((((((......))))))....))))...(((.(((((........))))))))))).)))) ( -36.80) >Bclau.0 2858068 120 - 4303871/2200-2320 GCCUUUAGUACGAUGCCGUAGACCCGAAACCGGGUGAUCUACCCAUGUCCAGGGUGAAGGUCGGGUAACACCGACUGGAGGCCCGAACCCACGCAAGUUGAAAAUUGCGGGGAUGAGGUG (((((((((.((.((((....(((((....)))))(((((((((.......))))..))))).))))....)))))))))))((..((((.(((((........)))))))).)..)).. ( -48.00) >Blich.0 3118231 119 + 4222334/2200-2320 G-CAUGAGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUG .-.......((((((..((((((((.......)).).)))))))))))((.((((...((((((......))))))....))))...(((.(((((........))))))))....)).. ( -42.50) >Bsubt.0 3172985 119 + 4214630/2200-2320 G-CAUGAGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUG .-.......((((((..((((((((.......)).).)))))))))))((.((((...((((((......))))))....))))...(((.(((((........))))))))....)).. ( -42.50) >Banth.0 5077122 118 - 5227293/2200-2320 G-UUU-AGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUG .-...-...((((((..((((((((.......)).).)))))))))))((.((((...((((((......))))))....))))...(((.(((((........))))))))....)).. ( -42.50) >Bcere.0 5074106 118 - 5224283/2200-2320 G-UUU-AGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUG .-...-...((((((..((((((((.......)).).)))))))))))((.((((...((((((......))))))....))))...(((.(((((........))))))))....)).. ( -42.50) >consensus G_CAU_AGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUG .........((((((..((((((((.......)).).)))))))))))((.((((...((((((......))))))....))))...(((.(((((........))))))))....)).. (-39.30 = -39.08 + -0.22)

| Location | 3,178,984 – 3,179,102 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.17 |
| Mean single sequence MFE | -42.52 |
| Consensus MFE | -35.47 |
| Energy contribution | -33.83 |
| Covariance contribution | -1.64 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 118 - 4202352/2280-2400 AAUGAACCGGCGAGUUACGAUGUCGUGCAAGGUUAAG-CUGAAAAAGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCG-AAAUAGUACGCCGUCGUAGACCCGAAACCGUGUGAUCU .....(((((((.(((((((((.(((((..((((..(-((.....)))..))))...((....))..((((.....)))).-.....))))).)))))).))).))...))).))..... ( -39.60) >Bclau.0 2858068 119 - 4303871/2280-2400 AAUGAACCGGCGAGUUACGGUAUCGUGCAAGGUUAAG-CUGAUGAAGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGCCUUUAGUACGAUGCCGUAGACCCGAAACCGGGUGAUCU ......((((((.(((((((((((((((..((((..(-((.....)))..))))((.((....))..((((.....)))))).....)))))))))))).))).))...))))....... ( -50.30) >Blich.0 3118231 119 + 4222334/2280-2400 AAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGUUUGAAAAGACGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCG-CAUGAGUACGUGGUCGUAGACCCGAAACCAGGUGAUCU .....(((((((.(((((((((.(((((..((((..((((....))))..))))((.((....))..((((.....)))))-)....))))).)))))).))).))...)).)))..... ( -42.90) >Bsubt.0 3172985 118 + 4214630/2280-2400 AAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAG-CAGAAGAUGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCG-CAUGAGUACGUGGUCGUAGACCCGAAACCAGGUGAUCU .....(((((((.(((((((((.(((((..((((..(-((.....)))..))))((.((....))..((((.....)))))-)....))))).)))))).))).))...)).)))..... ( -42.50) >Banth.0 5077122 117 - 5227293/2280-2400 AAUGAACUGGCGAGUUACGAUCCCGUGCGAGGUUAAG-CUGAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCG-UUU-AGUACGUGGUCGUAGACCCGAAACCAGGUGAUCU ......((((((.(((((((((.(((((..((((..(-((.....)))..))))((.((....))..((((.....)))))-)..-.))))).)))))).))).))...))))....... ( -40.60) >Bcere.0 5074106 117 - 5224283/2280-2400 AAUGAACCGGCGAGUUACGAUCCCGUGCGAGGUUAAG-CUGAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCG-UUU-AGUACGUGGUCGUAGACCCGAAACCAGGUGAUCU .....(((((((.(((((((((.(((((..((((..(-((.....)))..))))((.((....))..((((.....)))))-)..-.))))).)))))).))).))...)).)))..... ( -39.20) >consensus AAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAG_CUGAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCG_CAU_AGUACGUGGUCGUAGACCCGAAACCAGGUGAUCU .......(((....((((((((.(((((..((((..(.((.....)))..))))...((....))..((((.....)))).......))))).))))))))..))).............. (-35.47 = -33.83 + -1.64)

| Location | 3,178,984 – 3,179,103 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.91 |
| Mean single sequence MFE | -42.55 |
| Consensus MFE | -38.52 |
| Energy contribution | -37.58 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.977061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 119 - 4202352/2320-2440 AGUUGGAGCCCAUUAACGGGUGACAGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUGUCGUGCAAGGUUAAG-CUGAAAAAGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGC .((((..((((......))))..))))..(((((...((((........((..((.(((....)))))..((((..(-((.....)))..)))))).((....))...))))...))))) ( -42.10) >Bclau.0 2858068 119 - 4303871/2320-2440 AGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGGUAUCGUGCAAGGUUAAG-CUGAUGAAGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGC .((((..(((((....)))))..))))..(((((...((((((((((((........)))).))))((..((((..(-((.....)))..)))))).((....))...))))...))))) ( -46.50) >Blich.0 3118231 120 + 4222334/2320-2440 AGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGUUUGAAAAGACGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGC .((((..(((((....)))))..))))..(((((...((((......(((.((.......)))))(((..((((..((((....))))..)))))))((....))...))))...))))) ( -43.70) >Bsubt.0 3172985 118 + 4214630/2320-2440 AGUCAGAGCCCGUUAAC-GGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAG-CAGAAGAUGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGC .((((..(((.......-)))..))))..(((((...((((......(((.((.......)))))(((..((((..(-((.....)))..)))))))((....))...))))...))))) ( -36.30) >Banth.0 5077122 119 - 5227293/2320-2440 AGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACUGGCGAGUUACGAUCCCGUGCGAGGUUAAG-CUGAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGC .((((..(((((....)))))..))))..(((((...((((........((..((.(((....)))))..((((..(-((.....)))..)))))).((....))...))))...))))) ( -43.00) >Bcere.0 5074106 119 - 5224283/2320-2440 AGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCGAGGUUAAG-CUGAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGC ..((((((((((....)))))(.((((.((((((((.(((....((((..((...((((....)))))).))))...-)))))))))))..))))).((....))...)))))....... ( -43.70) >consensus AGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAG_CUGAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGC .((((..(((((....)))))..))))..(((((...((((........((..((.(((....)))))..((((..(.((.....)))..)))))).((....))...))))...))))) (-38.52 = -37.58 + -0.94)

| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.39 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -38.75 |
| Energy contribution | -38.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/2400-2520 UGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAAAGAUCCUGAAACCGUGUGCCUACAACUAGUUGGAGCCCAUUAACGGGUGACAGCGUGCCUUUUGUAG ..........((....))(((((((.....(((((((....))))((.((......)))).........))).........((((..((((......))))..))))..))))))).... ( -38.00) >Bclau.0 2858068 120 - 4303871/2400-2520 UGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAACUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAG ..........((....))(((((((.....(((((((....)))).((...((.....))...))....))).........((((..(((((....)))))..))))..))))))).... ( -40.60) >Blich.0 3118231 120 + 4222334/2400-2520 UGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAGUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAG ...(((....((....))....))).....(((((((....)))).((...((.....))...))....)))(((((((((((((..(((((....)))))..)))).)))...)))))) ( -41.30) >Bsubt.0 3172985 119 + 4214630/2400-2520 UGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAGUAGUCAGAGCCCGUUAAC-GGUGAUGGCGUGCCUUUUGUAG ....(((.((((((..(((...........((.((((....)))).))..........((((.....(((....))).....))))..)))....))-)))).))).............. ( -34.80) >Banth.0 5077122 120 - 5227293/2400-2520 UGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAG ...(((....((....))....))).....(((((((....)))).((...((.....))...))....)))(((((((..((((..(((((....)))))..))))......))))))) ( -40.90) >Bcere.0 5074106 120 - 5224283/2400-2520 UGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAG ...(((....((....))....))).....(((((((....)))).((...((.....))...))....)))(((((((..((((..(((((....)))))..))))......))))))) ( -40.90) >consensus UGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAG .......((((((...((((..(.(....).).((((....))))...........)))).....)).))))(((((((..((((..(((((....)))))..))))......))))))) (-38.75 = -38.75 + 0.00)

| Location | 3,178,984 – 3,179,101 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -22.75 |
| Energy contribution | -21.65 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 117 - 4202352/2600-2720 CAGGAUCCACUCUGGAGGAAACGAAGUUUCGGCUACAGGGUUGUUACCU-UCUGUGACGGACC-UUUCCAGGUCGCUUCACCUACUUCGUUUCUUUCUGACUCCGU-AUAGAGUGUCCUA .((((..((((((((((((((((((((...(((((((((((....))).-.)))))...((((-(....))))))))......)))))))))))))).(....)..-..)))))))))). ( -42.10) >Bclau.0 2858068 117 - 4303871/2600-2720 CAGGAUCCACUCUGGAGGAAACGAAGUUUCAGCUACAGGGCUGUUACCU-UCUUCGGCCUGCC-UUUCCAGACAG-UUCACCUACUCCGUUUCUUGAUCACUCCGUUGUUGAGUGUCCUA .((((..((((((((((((((((((((..(((((....)))))..).))-))...(..(((.(-(....)).)))-..).........))))).......))))).....))))))))). ( -31.31) >Blich.0 3118231 119 + 4222334/2600-2720 CAGGAUCCACUCAGGAGAGAACGAAGUUUUGACUACAGGGCUGUUACCUUCCUAUGGCGGGCC-UUUCCAGACCUCUUCAUCUACCUCGUUCUUUUGUAACUCCGGUACAGAGUGUCCUA .((((..(((((..((((((((((.((..(((....(((.((((((........)))))).))-)....((....)))))...)).))))))))))(((.(....)))).))))))))). ( -33.90) >Bsubt.0 3172985 118 + 4214630/2600-2720 CAGGAUCCACUCAGGAGAGAACGAAGUUUUGACUACAGGGCUGUUACCU-CCUAUGGCGGGCCCUUUCCAGACCUCUUCAUCUACCUCGUUCUUUUGUAACUCCG-UACAGAGUGUCCUA .((((..(((((..((((((((((.((..(((....((((((....((.-.....))..))))))....((....)))))...)).))))))))))(((......-))).))))))))). ( -35.10) >Banth.0 5077122 117 - 5227293/2600-2720 CAGGAUACAUUCAAGAGAGAACGAAGUUUCGACUACGGGGUUGUUACCC-UCUACGACGGACC-UUUCCAGGUCGCUUCGUCUACCUCGUUCCUUUGUAACUCCG-UAUAGAAUGUCCUA .((((..(((((.((((.((((((.((...(((...(((((....))))-)....((.(((((-(....))))).).))))).)).))))))))))(((......-))).))))))))). ( -34.30) >Bcere.0 5074106 117 - 5224283/2600-2720 CAGGAUACAUUCAAGAGAGAACGAAGUUUCGACUACGGGGUUGUUACCC-UCUACGACGGACC-UUUCCAGGUCGCUUCGUCUACCUCGUUCCUUUGUAACUCCG-UAUAGAAUGUCCUA .((((..(((((.((((.((((((.((...(((...(((((....))))-)....((.(((((-(....))))).).))))).)).))))))))))(((......-))).))))))))). ( -34.30) >consensus CAGGAUCCACUCAGGAGAGAACGAAGUUUCGACUACAGGGCUGUUACCU_UCUACGACGGACC_UUUCCAGACCGCUUCAUCUACCUCGUUCCUUUGUAACUCCG_UAUAGAGUGUCCUA .((((..(((((...(((((((((.((..(((....(((.......))).........(((.....)))........)))...)).)))))))))((((.......))))))))))))). (-22.75 = -21.65 + -1.10)
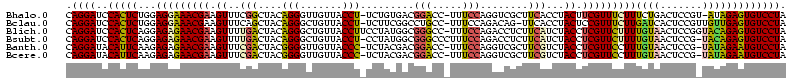
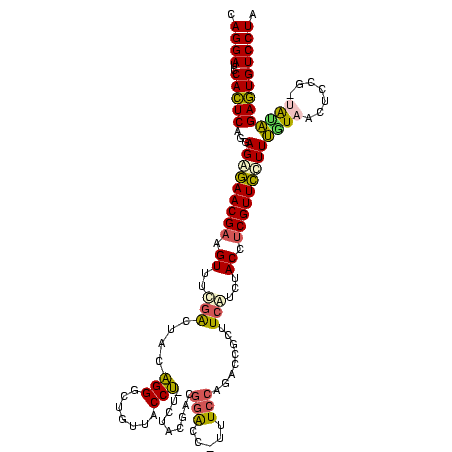
| Location | 3,178,984 – 3,179,101 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -41.38 |
| Consensus MFE | -35.86 |
| Energy contribution | -31.68 |
| Covariance contribution | -4.19 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 117 - 4202352/2600-2720 UAGGACACUCUAU-ACGGAGUCAGAAAGAAACGAAGUAGGUGAAGCGACCUGGAAA-GGUCCGUCACAGA-AGGUAACAACCCUGUAGCCGAAACUUCGUUUCCUCCAGAGUGGAUCCUG (((((((((((..-..((((.......((((((((((..((((.(.(((((....)-))))).))))...-.(((.(((....))).)))...))))))))))))))))))))..))))) ( -46.61) >Bclau.0 2858068 117 - 4303871/2600-2720 UAGGACACUCAACAACGGAGUGAUCAAGAAACGGAGUAGGUGAA-CUGUCUGGAAA-GGCAGGCCGAAGA-AGGUAACAGCCCUGUAGCUGAAACUUCGUUUCCUCCAGAGUGGAUCCUG ((((((((((.......((....))..((((((((((.......-((((((....)-)))))(((.....-.)))..((((......))))..)))))))))).....)))))..))))) ( -42.00) >Blich.0 3118231 119 + 4222334/2600-2720 UAGGACACUCUGUACCGGAGUUACAAAAGAACGAGGUAGAUGAAGAGGUCUGGAAA-GGCCCGCCAUAGGAAGGUAACAGCCCUGUAGUCAAAACUUCGUUCUCUCCUGAGUGGAUCCUG ((((((((((......((((.......((((((((((.(((..((.((.(((....-.(((..((...))..)))..)))))))...)))...)))))))))))))).)))))..))))) ( -37.91) >Bsubt.0 3172985 118 + 4214630/2600-2720 UAGGACACUCUGUA-CGGAGUUACAAAAGAACGAGGUAGAUGAAGAGGUCUGGAAAGGGCCCGCCAUAGG-AGGUAACAGCCCUGUAGUCAAAACUUCGUUCUCUCCUGAGUGGAUCCUG ((((((((((....-.((((.......((((((((((((((......)))).((.(((((..(((.....-.)))....)))))....))...)))))))))))))).)))))..))))) ( -39.51) >Banth.0 5077122 117 - 5227293/2600-2720 UAGGACAUUCUAUA-CGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAA-GGUCCGUCGUAGA-GGGUAACAACCCCGUAGUCGAAACUUCGUUCUCUCUUGAAUGUAUCCUG ((((((((((....-.((((.......((((((((((.(((((.(.(((((....)-))))).))...(.-((((....)))))...)))...)))))))))))))).)))))..))))) ( -41.11) >Bcere.0 5074106 117 - 5224283/2600-2720 UAGGACAUUCUAUA-CGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAA-GGUCCGUCGUAGA-GGGUAACAACCCCGUAGUCGAAACUUCGUUCUCUCUUGAAUGUAUCCUG ((((((((((....-.((((.......((((((((((.(((((.(.(((((....)-))))).))...(.-((((....)))))...)))...)))))))))))))).)))))..))))) ( -41.11) >consensus UAGGACACUCUAUA_CGGAGUUACAAAGGAACGAGGUAGAUGAAGCGACCUGGAAA_GGCCCGCCAUAGA_AGGUAACAACCCUGUAGUCGAAACUUCGUUCUCUCCUGAGUGGAUCCUG ((((((((((......((((.......((((((((((.(((.....(((((....).))))..........(((.......)))...)))...)))))))))))))).)))))..))))) (-35.86 = -31.68 + -4.19)
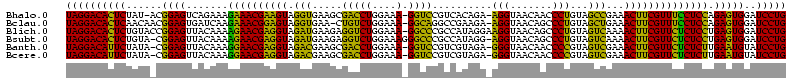
| Location | 3,178,984 – 3,179,100 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.07 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -26.72 |
| Energy contribution | -26.00 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 116 - 4202352/2640-2760 GGAA-ACAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACACUCUAU-ACGGAGUCAGAAAGAAACGAAGUAGGUGAAGCGACCUGGAAA-GGUCCGUCACAGA-AGGUAACAA ....-(((((((....(((((((....))))))))))))))...((..(((..-.((...((.....))..))......((((.(.(((((....)-))))).)))))))-..))..... ( -38.50) >Bclau.0 2858068 116 - 4303871/2640-2760 GGAA-ACAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACACUCAACAACGGAGUGAUCAAGAAACGGAGUAGGUGAA-CUGUCUGGAAA-GGCAGGCCGAAGA-AGGUAACAG (...-.).((((...((((((((....)))))))).(((....(((((((.......))))).))....))))).)).......-((((((....)-)))))(((.....-.)))..... ( -41.20) >Blich.0 3118231 119 + 4222334/2640-2760 GGGGAACAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACACUCUGUACCGGAGUUACAAAAGAACGAGGUAGAUGAAGAGGUCUGGAAA-GGCCCGCCAUAGGAAGGUAACAG (((..(((((((....(((((((....)))))))))))))).((((.((((.((((...(((.......)))..)))).....)))))))).....-..)))(((.......)))..... ( -41.20) >Bsubt.0 3172985 117 + 4214630/2640-2760 CGGGAUCAGCCCAAACCAAGAGGCUUGCCUC-UGUGGUUGUAGGACACUCUGUA-CGGAGUUACAAAAGAACGAGGUAGAUGAAGAGGUCUGGAAAGGGCCCGCCAUAGG-AGGUAACAG .(..(((.((((.((((((((((....))))-).)))))...((((.((((.((-(...(((.......)))...))).....)))))))).....))))((......))-.)))..).. ( -35.10) >Banth.0 5077122 117 - 5227293/2640-2760 GGAACAUAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACAUUCUAUA-CGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAA-GGUCCGUCGUAGA-GGGUAACAA ........((((...((((((((....))))))))...(.((.(((..(((((.-....(((.......)))...)))))....(.(((((....)-)))))))).)).)-))))..... ( -39.00) >Bcere.0 5074106 117 - 5224283/2640-2760 GGAACAUAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACAUUCUAUA-CGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAA-GGUCCGUCGUAGA-GGGUAACAA ........((((...((((((((....))))))))...(.((.(((..(((((.-....(((.......)))...)))))....(.(((((....)-)))))))).)).)-))))..... ( -39.00) >consensus GGAA_ACAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACACUCUAUA_CGGAGUUACAAAGGAACGAGGUAGAUGAAGCGACCUGGAAA_GGCCCGCCAUAGA_AGGUAACAA .....(((((((....(((((((....)))))))))))))).....(((((.....))))).................((((....(((((....).))))))))............... (-26.72 = -26.00 + -0.72)

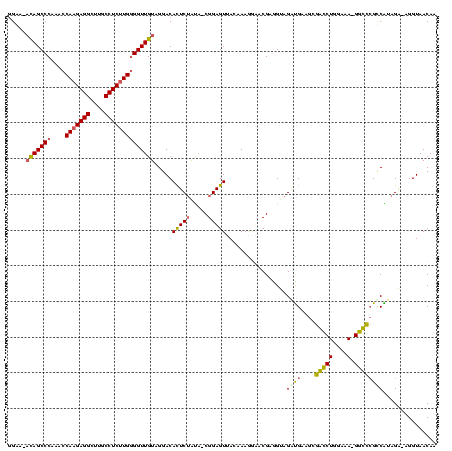
| Location | 3,178,984 – 3,179,102 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.57 |
| Mean single sequence MFE | -36.56 |
| Consensus MFE | -32.39 |
| Energy contribution | -31.45 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 118 - 4202352/2720-2840 AGACCUGGGGAACUGAAACAUCUAAGUACCCAGAGGAAGAGAAAGCAAAUGCGAUUUCC-UGAGUAGCGGCGAGCGAAACGGAA-ACAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUG .((((((((...........((....(((.(((.((((......((....))..)))))-)).)))((.....))))...(...-.)..))))..((((((((....)))))))))))). ( -36.20) >Bclau.0 2858068 118 - 4303871/2720-2840 AGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGAAGAGAAAGCAAAUGCGAUUUCC-UGAGUAGCGGCGAGCGAAACGGAA-ACAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUG ....(((((..(((..........))).)))))((((((.....((....))..)))))-).....((.....)).........-.((((((....(((((((....))))))))))))) ( -37.60) >Blich.0 3118231 120 + 4222334/2720-2840 AGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGAAGAGAAAGCAAAUGCGAUUCCCUUGAGUAGCGGCGAGCGAAACGGGGAACAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUG ....(((((..(((..........))).)))))...........((....))..((((((......((.....)).....))))))((((((....(((((((....))))))))))))) ( -40.60) >Bsubt.0 3172985 116 + 4214630/2720-2840 AGACCCGGGGAACUGAAACAUCUAAGUACCCGGA-GAAGAGAAAGCAAAUGCGAUUCCC-UGAGUAGCGGCGA-CGAACACGGGAUCAGCCCAAACCAAGAGGCUUGCCUC-UGUGGUUG ....(((((..(((..........))).))))).-.........((....))((((((.-((.....((....-))..)).))))))......((((((((((....))))-).))))). ( -36.10) >Banth.0 5077122 119 - 5227293/2720-2840 AGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGGAAGAGAAAGCAAAUGCGAUUUCC-UGAGUAGCGGCGAGCGAAACGGAACAUAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUG ....(((((..(((..........))).)))))((((((.....((....))..)))))-)...(((((((.................)))....((((((((....)))))))).)))) ( -34.43) >Bcere.0 5074106 119 - 5224283/2720-2840 AGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGGAAGAGAAAGCAAAUGCGAUUUCC-UGAGUAGCGGCGAGCGAAACGGAACAUAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUG ....(((((..(((..........))).)))))((((((.....((....))..)))))-)...(((((((.................)))....((((((((....)))))))).)))) ( -34.43) >consensus AGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGAAGAGAAAGCAAAUGCGAUUUCC_UGAGUAGCGGCGAGCGAAACGGAA_ACAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUG ....(((((..(((..........))).)))))...........((....))...((((.......((.....)).....))))..((((((....(((((((....))))))))))))) (-32.39 = -31.45 + -0.94)
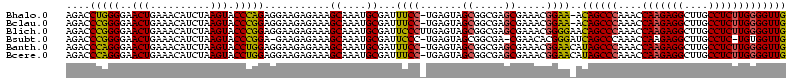
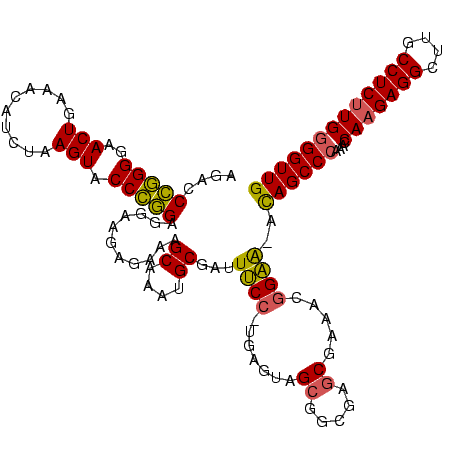
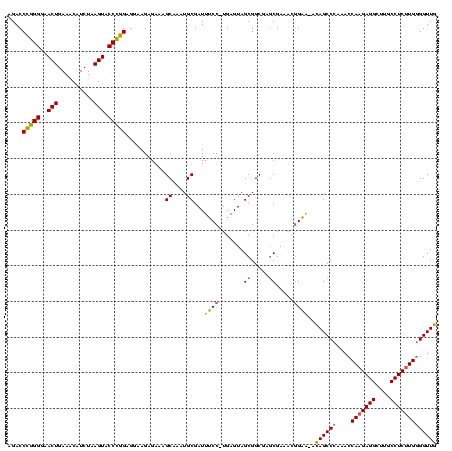
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.88 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -34.06 |
| Energy contribution | -30.35 |
| Covariance contribution | -3.71 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/2800-2920 UCUUCCUCUGGGUACUUAGAUGUUUCAGUUCCCCAGGUCUGCCUCCUCAUACCCUAUAGAUUCAGGUAUGGGUACCAUCCCAUUACGGAUGGUGGGUUCCCCCAUUCGGAUAUCCCCGGA ...(((.(((((.(((..((....)))))..)))))(((((....((((((((...........)))))))).(((((((......)))))))(((....)))...)))))......))) ( -37.20) >Bclau.0 2858068 120 - 4303871/2800-2920 UCUUCCUCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUCUCGACAUCCUAUGUGUUCAGAUGCCAGUACCAUCCCAUUACGGAUGGUGGGUUCCCCCAUUCGGAAAUCUCCGGA ......((((((.(((..((....)))))..)))))).((((.(((.(((((.....))))).))).).))).(((((((......)))))))(((....))).((((((....)))))) ( -37.60) >Blich.0 3118231 120 + 4222334/2800-2920 UCUUCCUCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACUGCUCCAUUACGAGCAGUGGGUUUCCCCAUUCGGAAAUCUCCGGA ...(((.(((((.(((..((....)))))..)))))(((((..(((...........)))..)))))..(((((((((((......))))))))(((((((......))))))))))))) ( -39.20) >Bsubt.0 3172985 119 + 4214630/2800-2920 UCUUC-UCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACCACUCCAUUACGAGUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGA .....-.(((((.(((..((....)))))..)))))(((((..(((...........)))..))))).((((((((((((......))))))))(((((((......))))))))))).. ( -37.60) >Banth.0 5077122 119 - 5227293/2800-2920 UCUUCCUCCAGGUACUUAGAUGUUUCAGUUCCCUGGGUCUGUCUUC-CUUACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGA ...(((((((((.(((..((....)))))..)))))(((((.....-..(((.....)))..))))).))))((((((((......))))))))(((((((......)))))))...... ( -31.50) >Bcere.0 5074106 119 - 5224283/2800-2920 UCUUCCUCCAGGUACUUAGAUGUUUCAGUUCCCUGGGUCUGUCUUC-CUUACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGA ...(((((((((.(((..((....)))))..)))))(((((.....-..(((.....)))..))))).))))((((((((......))))))))(((((((......)))))))...... ( -31.50) >consensus UCUUCCUCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUCUCAUACCCUAUGUAUUCAGAUAUGGAUACCAUACCAUUACGAAUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGA ......((((((.(((..((....)))))..)))))).......((.((((((...........)))))))).(((((((......)))))))(((....))).((((((....)))))) (-34.06 = -30.35 + -3.71)
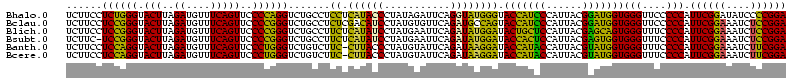
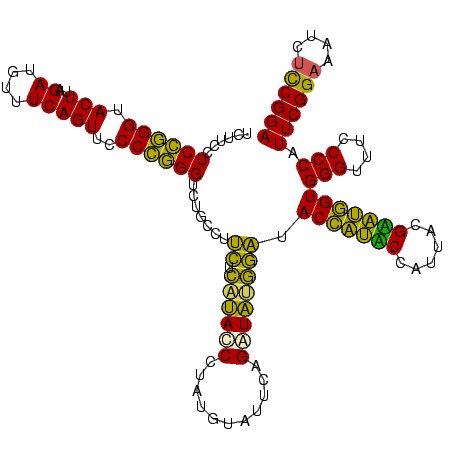
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.88 |
| Mean single sequence MFE | -41.88 |
| Consensus MFE | -43.19 |
| Energy contribution | -38.95 |
| Covariance contribution | -4.24 |
| Combinations/Pair | 1.47 |
| Mean z-score | -3.54 |
| Structure conservation index | 1.03 |
| SVM decision value | 4.56 |
| SVM RNA-class probability | 0.999920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/2800-2920 UCCGGGGAUAUCCGAAUGGGGGAACCCACCAUCCGUAAUGGGAUGGUACCCAUACCUGAAUCUAUAGGGUAUGAGGAGGCAGACCUGGGGAACUGAAACAUCUAAGUACCCAGAGGAAGA ..((((....))))...(((....)))(((((((......))))))).(((((((((.........))))))).)).......(((.(((.(((..........))).)))..))).... ( -42.60) >Bclau.0 2858068 120 - 4303871/2800-2920 UCCGGAGAUUUCCGAAUGGGGGAACCCACCAUCCGUAAUGGGAUGGUACUGGCAUCUGAACACAUAGGAUGUCGAGAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGAAGA (((..((((..(((...(((....)))(((((((......)))))))..))).)))).........(..((((....))))..)(((((..(((..........))).))))).)))... ( -42.20) >Blich.0 3118231 120 + 4222334/2800-2920 UCCGGAGAUUUCCGAAUGGGGAAACCCACUGCUCGUAAUGGAGCAGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGAAGA ..(((......)))...(((....)))(((((((......))))))).(((...((((..(((.((.....)).)))..)))).(((((..(((..........))).))))).)))... ( -43.70) >Bsubt.0 3172985 119 + 4214630/2800-2920 UCCGGAGAUUUCCGAAUGGGGAAACCCACCACUCGUAAUGGAGUGGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGA-GAAGA ..(((......)))...(((....)))(((((((......))))))).(((((((((.........))))))).))........(((((..(((..........))).))))).-..... ( -41.00) >Banth.0 5077122 119 - 5227293/2800-2920 UCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUAAG-GAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGGAAGA ........(((((....(((....)))(((((((......))))))).(((((((((.........)))))))-))........(((((..(((..........))).))))).))))). ( -40.90) >Bcere.0 5074106 119 - 5224283/2800-2920 UCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUAAG-GAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGGAAGA ........(((((....(((....)))(((((((......))))))).(((((((((.........)))))))-))........(((((..(((..........))).))))).))))). ( -40.90) >consensus UCCGGAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGAAUGGUAUCCAUAUCUGAAUACAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGAAGA ........(((((....(((....)))(((((((......))))))).(((((((((.........))))))).))........(((((..(((..........))).))))).))))). (-43.19 = -38.95 + -4.24)
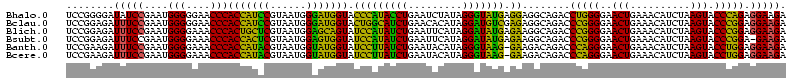
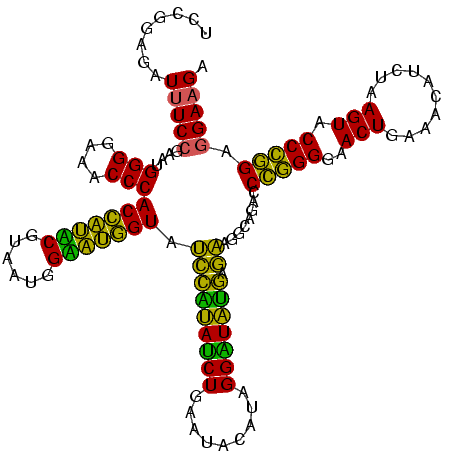
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.33 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -28.46 |
| Energy contribution | -25.80 |
| Covariance contribution | -2.66 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/2840-2960 GCCUCCUCAUACCCUAUAGAUUCAGGUAUGGGUACCAUCCCAUUACGGAUGGUGGGUUCCCCCAUUCGGAUAUCCCCGGAUCAAAGCUUACUUACAGCUCCCCGAGGCGUUUCGCCGUUC (((((((((((((...........)))))))).(((((((......)))))))(((.......((((((......))))))...((((.......)))).))))))))............ ( -40.10) >Bclau.0 2858068 120 - 4303871/2840-2960 GCCUCUCGACAUCCUAUGUGUUCAGAUGCCAGUACCAUCCCAUUACGGAUGGUGGGUUCCCCCAUUCGGAAAUCUCCGGAUCGAAGCUUACUUACAGCUCCCCGAAGCCUAUCGCGGUUC ((.(((.(((((.....))))).))).))....(((((((......)))))))(((....))).((((((....))))))(((.((((.......))))...)))((((......)))). ( -33.60) >Blich.0 3118231 120 + 4222334/2840-2960 GCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACUGCUCCAUUACGAGCAGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACAGCUCCCCGAAGCAUAUCGGUGUUC (((...(((((...))))).....((((((...(((((((......)))))))(((.......(((((((....)))))))...((((.......)))).)))....))))))))).... ( -34.10) >Bsubt.0 3172985 120 + 4214630/2840-2960 GCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACCACUCCAUUACGAGUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACAGCUCCCCGAAGCAUAUCGGUGUUC (((...(((((...))))).....((((((...(((((((......)))))))(((.......(((((((....)))))))...((((.......)))).)))....))))))))).... ( -34.10) >Banth.0 5077122 119 - 5227293/2840-2960 GUCUUC-CUUACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACAGCUCCCCGAAGCAUAUCGGCGUUA (((.((-((((..((........)).))))))((((((((......))))))))(((((((......)))))))(((((.....((((.......))))..))))).......))).... ( -29.60) >Bcere.0 5074106 119 - 5224283/2840-2960 GUCUUC-CUUACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACAGCUCCCCGAAGCAUAUCGGCGUUA (((.((-((((..((........)).))))))((((((((......))))))))(((((((......)))))))(((((.....((((.......))))..))))).......))).... ( -29.60) >consensus GCCUUCUCAUACCCUAUGUAUUCAGAUAUGGAUACCAUACCAUUACGAAUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUACUUACAGCUCCCCGAAGCAUAUCGGCGUUC ....((.((((((...........)))))))).(((((((......)))))))(((....)))(((((((....)))))))...((((.......))))..((((......))))..... (-28.46 = -25.80 + -2.66)

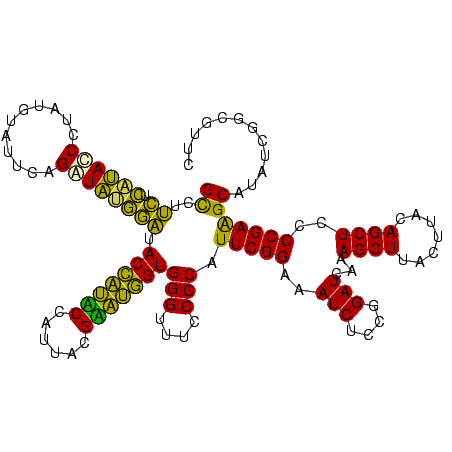
| Location | 3,178,984 – 3,179,104 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.33 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -45.02 |
| Energy contribution | -40.67 |
| Covariance contribution | -4.35 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.99 |
| Structure conservation index | 1.05 |
| SVM decision value | 4.04 |
| SVM RNA-class probability | 0.999769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3178984 120 - 4202352/2840-2960 GAACGGCGAAACGCCUCGGGGAGCUGUAAGUAAGCUUUGAUCCGGGGAUAUCCGAAUGGGGGAACCCACCAUCCGUAAUGGGAUGGUACCCAUACCUGAAUCUAUAGGGUAUGAGGAGGC ...(((.......((((((((((((.......)))))...)))))))....)))...(((....)))(((((((......))))))).(((((((((.........))))))).)).... ( -47.20) >Bclau.0 2858068 120 - 4303871/2840-2960 GAACCGCGAUAGGCUUCGGGGAGCUGUAAGUAAGCUUCGAUCCGGAGAUUUCCGAAUGGGGGAACCCACCAUCCGUAAUGGGAUGGUACUGGCAUCUGAACACAUAGGAUGUCGAGAGGC ....((.((....((((((((((((.......))))))...))))))...))))...(((....)))(((((((......))))))).(((((((((.........))))))).)).... ( -43.20) >Blich.0 3118231 120 + 4222334/2840-2960 GAACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACUGCUCGUAAUGGAGCAGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGC .....((((((((((((((((((((.......)))))...)))))))..........(((....)))(((((((......)))))))...))))))....((((((...))))))..)). ( -41.30) >Bsubt.0 3172985 120 + 4214630/2840-2960 GAACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACCACUCGUAAUGGAGUGGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGC .....((((((((((((((((((((.......)))))...)))))))..........(((....)))(((((((......)))))))...))))))....((((((...))))))..)). ( -41.30) >Banth.0 5077122 119 - 5227293/2840-2960 UAACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUAAG-GAAGAC ......((.....((((((((((((.......)))))...))))))).....))...(((....)))(((((((......))))))).(((((((((.........)))))))-)).... ( -42.20) >Bcere.0 5074106 119 - 5224283/2840-2960 UAACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUAAG-GAAGAC ......((.....((((((((((((.......)))))...))))))).....))...(((....)))(((((((......))))))).(((((((((.........)))))))-)).... ( -42.20) >consensus GAACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGAAUGGUAUCCAUAUCUGAAUACAUAGGAUAUGAGAAGGC ......((.....((((((((((((.......)))))...))))))).....))...(((....)))(((((((......))))))).(((((((((.........))))))).)).... (-45.02 = -40.67 + -4.35)

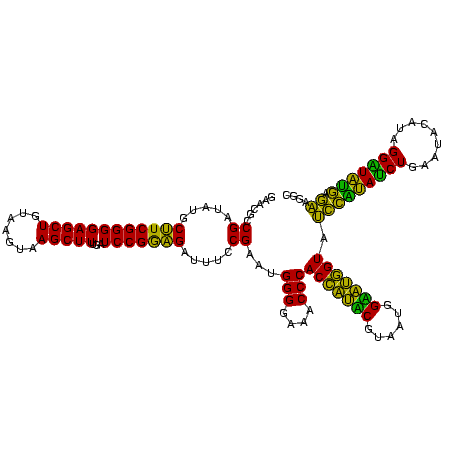
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:19:11 2006