| Sequence ID | Bsubt.0 |
|---|---|
| Location | 2,607,929 – 2,608,022 |
| Length | 93 |
| Max. P | 0.999982 |

| Location | 2,607,929 – 2,608,022 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 67.61 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -21.77 |
| Energy contribution | -23.90 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.18 |
| Mean z-score | -4.29 |
| Structure conservation index | 0.65 |
| SVM decision value | 5.28 |
| SVM RNA-class probability | 0.999982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
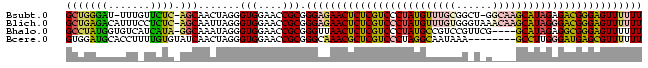
>Bsubt.0 2607929 93 + 4214630 AAAAAACUCCCGUCUCUAUGCUUGCC-AGCCGCAAACAUAGGGACGAGAGUUCUCCCGCGGUUCCACCCUAGUUGCU-GAGAACAAA-AUCCCAGC ....(((((.((((((((((.((((.-....)))).)))))))))).))))).......((.....))......(((-(.((.....-.)).)))) ( -30.30) >Blich.0 2615034 95 + 4222334 AAAAAACUCCCGUCCCUAUGCUUGUUUACCCACAAACAUAGGGACGAGAGUUCUCCCGCGGUUCCACCCUAAUUGCU-GAGAGGAAAUGUCUCAGC ....(((((.((((((((((.((((......)))).)))))))))).))))).......((.....))......(((-((((.......))))))) ( -35.70) >Bhalo.0 2748379 91 - 4202352 AAAAAACUCCCGCCUCUAUGC----CGAACGGACGGCAUAGGGACGAGAGUUAACCCGCGGUUCCACCCUAUUUGCC-UAUGAUGACACCAUAGGC ....(((((.((.((((((((----((......)))))))))).)).))))).......((.....))......(((-((((.......))))))) ( -34.40) >Bcere.0 552434 88 - 5224283 AAAAAACGCUCAUCCCAAGGC--------UUUAUUGCCUAGGGACGAGCGUUUGCCCGCGGUUCCACCCUAGUUGAUACACAAAAGGUGCAUCCAC ...((((((((.((((.((((--------......)))).)))).))))))))......((...((((....(((.....)))..))))...)).. ( -34.50) >consensus AAAAAACUCCCGUCCCUAUGC____C_A_CCGAAAACAUAGGGACGAGAGUUCUCCCGCGGUUCCACCCUAGUUGCU_GAGAAAAAAUGCAUCAGC ....(((((.(((((((((((((((......))))))))))))))).))))).......((.....))............................ (-21.77 = -23.90 + 2.13)


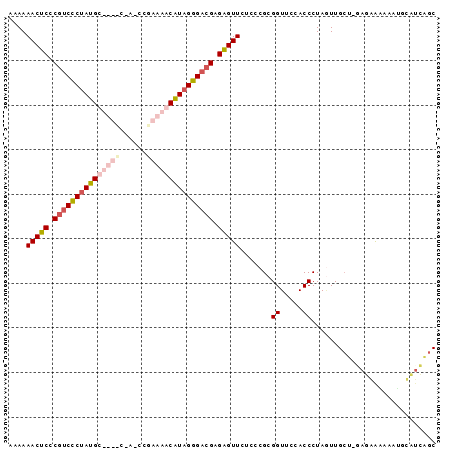
| Location | 2,607,929 – 2,608,022 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 67.61 |
| Mean single sequence MFE | -47.20 |
| Consensus MFE | -36.92 |
| Energy contribution | -37.68 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.43 |
| Mean z-score | -6.37 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 2607929 93 + 4214630 GCUGGGAU-UUUGUUCUC-AGCAACUAGGGUGGAACCGCGGGAGAACUCUCGUCCCUAUGUUUGCGGCU-GGCAAGCAUAGAGACGGGAGUUUUUU (((((((.-.....))))-))).......(((....))).((((((((((((((.((((((((((....-.)))))))))).)))))))))))))) ( -47.50) >Blich.0 2615034 95 + 4222334 GCUGAGACAUUUCCUCUC-AGCAAUUAGGGUGGAACCGCGGGAGAACUCUCGUCCCUAUGUUUGUGGGUAAACAAGCAUAGGGACGGGAGUUUUUU (((((((.......))))-))).......(((....))).(((((((((((((((((((((((((......))))))))))))))))))))))))) ( -52.70) >Bhalo.0 2748379 91 - 4202352 GCCUAUGGUGUCAUCAUA-GGCAAAUAGGGUGGAACCGCGGGUUAACUCUCGUCCCUAUGCCGUCCGUUCG----GCAUAGAGGCGGGAGUUUUUU (((((((((...))))))-))).......(((....))).....((((((((((.((((((((......))----)))))).)))))))))).... ( -43.50) >Bcere.0 552434 88 - 5224283 GUGGAUGCACCUUUUGUGUAUCAACUAGGGUGGAACCGCGGGCAAACGCUCGUCCCUAGGCAAUAAA--------GCCUUGGGAUGAGCGUUUUUU ((((.(.((((((..((......)).)))))).).))))....(((((((((((((.((((......--------)))).)))))))))))))... ( -45.10) >consensus GCUGAGACAUUUAUUCUC_AGCAACUAGGGUGGAACCGCGGGAGAACUCUCGUCCCUAUGCUUGCGG_U_G____GCAUAGAGACGGGAGUUUUUU (((((((.......)))).))).......(((....))).(((((((((((((((((((((((((......))))))))))))))))))))))))) (-36.92 = -37.68 + 0.75)
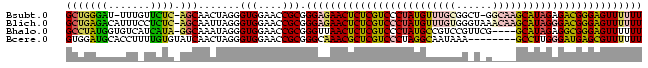
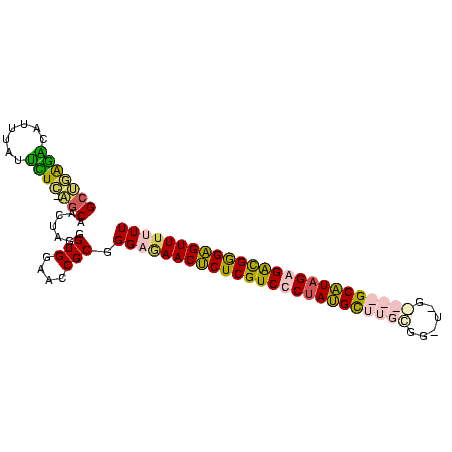
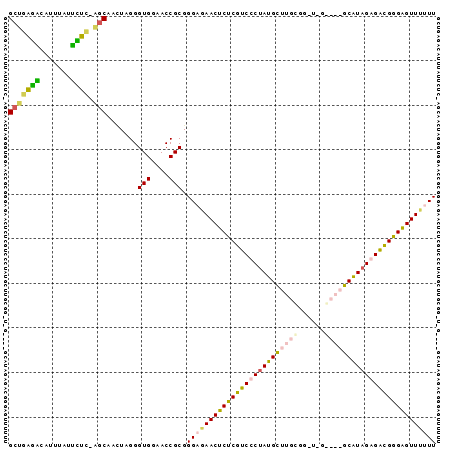
| Location | 2,607,929 – 2,608,022 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 67.61 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -21.77 |
| Energy contribution | -23.90 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.18 |
| Mean z-score | -4.29 |
| Structure conservation index | 0.65 |
| SVM decision value | 5.28 |
| SVM RNA-class probability | 0.999982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 2607929 93 + 4214630/0-96 AAAAAACUCCCGUCUCUAUGCUUGCC-AGCCGCAAACAUAGGGACGAGAGUUCUCCCGCGGUUCCACCCUAGUUGCU-GAGAACAAA-AUCCCAGC ....(((((.((((((((((.((((.-....)))).)))))))))).))))).......((.....))......(((-(.((.....-.)).)))) ( -30.30) >Blich.0 2615034 95 + 4222334/0-96 AAAAAACUCCCGUCCCUAUGCUUGUUUACCCACAAACAUAGGGACGAGAGUUCUCCCGCGGUUCCACCCUAAUUGCU-GAGAGGAAAUGUCUCAGC ....(((((.((((((((((.((((......)))).)))))))))).))))).......((.....))......(((-((((.......))))))) ( -35.70) >Bhalo.0 2748379 91 - 4202352/0-96 AAAAAACUCCCGCCUCUAUGC----CGAACGGACGGCAUAGGGACGAGAGUUAACCCGCGGUUCCACCCUAUUUGCC-UAUGAUGACACCAUAGGC ....(((((.((.((((((((----((......)))))))))).)).))))).......((.....))......(((-((((.......))))))) ( -34.40) >Bcere.0 552434 88 - 5224283/0-96 AAAAAACGCUCAUCCCAAGGC--------UUUAUUGCCUAGGGACGAGCGUUUGCCCGCGGUUCCACCCUAGUUGAUACACAAAAGGUGCAUCCAC ...((((((((.((((.((((--------......)))).)))).))))))))......((...((((....(((.....)))..))))...)).. ( -34.50) >consensus AAAAAACUCCCGUCCCUAUGC____C_A_CCGAAAACAUAGGGACGAGAGUUCUCCCGCGGUUCCACCCUAGUUGCU_GAGAAAAAAUGCAUCAGC ....(((((.(((((((((((((((......))))))))))))))).))))).......((.....))............................ (-21.77 = -23.90 + 2.13)

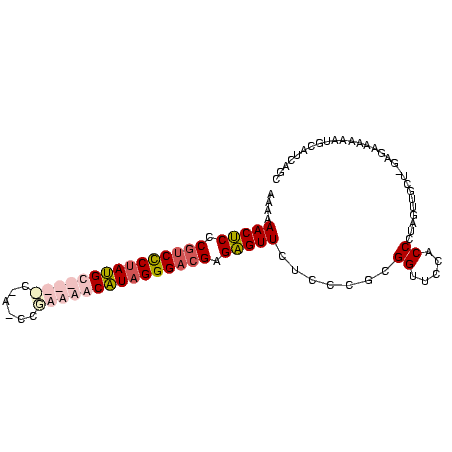
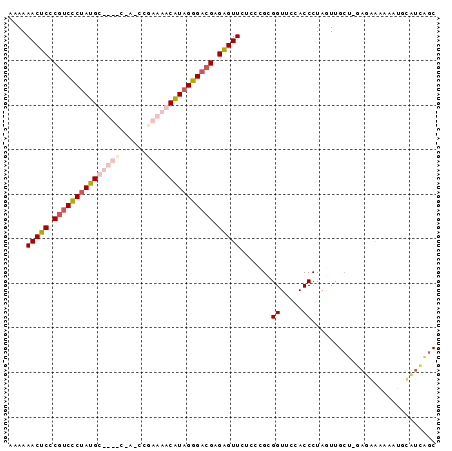
| Location | 2,607,929 – 2,608,022 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 67.61 |
| Mean single sequence MFE | -47.20 |
| Consensus MFE | -36.92 |
| Energy contribution | -37.68 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.43 |
| Mean z-score | -6.37 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 2607929 93 + 4214630/0-96 GCUGGGAU-UUUGUUCUC-AGCAACUAGGGUGGAACCGCGGGAGAACUCUCGUCCCUAUGUUUGCGGCU-GGCAAGCAUAGAGACGGGAGUUUUUU (((((((.-.....))))-))).......(((....))).((((((((((((((.((((((((((....-.)))))))))).)))))))))))))) ( -47.50) >Blich.0 2615034 95 + 4222334/0-96 GCUGAGACAUUUCCUCUC-AGCAAUUAGGGUGGAACCGCGGGAGAACUCUCGUCCCUAUGUUUGUGGGUAAACAAGCAUAGGGACGGGAGUUUUUU (((((((.......))))-))).......(((....))).(((((((((((((((((((((((((......))))))))))))))))))))))))) ( -52.70) >Bhalo.0 2748379 91 - 4202352/0-96 GCCUAUGGUGUCAUCAUA-GGCAAAUAGGGUGGAACCGCGGGUUAACUCUCGUCCCUAUGCCGUCCGUUCG----GCAUAGAGGCGGGAGUUUUUU (((((((((...))))))-))).......(((....))).....((((((((((.((((((((......))----)))))).)))))))))).... ( -43.50) >Bcere.0 552434 88 - 5224283/0-96 GUGGAUGCACCUUUUGUGUAUCAACUAGGGUGGAACCGCGGGCAAACGCUCGUCCCUAGGCAAUAAA--------GCCUUGGGAUGAGCGUUUUUU ((((.(.((((((..((......)).)))))).).))))....(((((((((((((.((((......--------)))).)))))))))))))... ( -45.10) >consensus GCUGAGACAUUUAUUCUC_AGCAACUAGGGUGGAACCGCGGGAGAACUCUCGUCCCUAUGCUUGCGG_U_G____GCAUAGAGACGGGAGUUUUUU (((((((.......)))).))).......(((....))).(((((((((((((((((((((((((......))))))))))))))))))))))))) (-36.92 = -37.68 + 0.75)
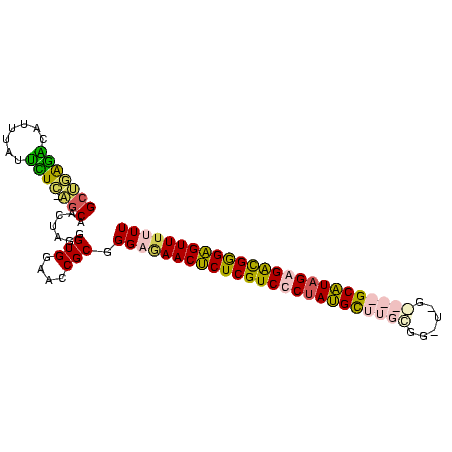
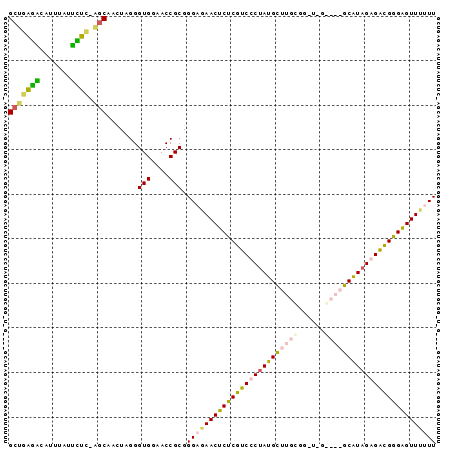
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:17:31 2006