
| Sequence ID | Bsubt.0 |
|---|---|
| Location | 2,548,606 – 2,548,714 |
| Length | 108 |
| Max. P | 0.999999 |

| Location | 2,548,606 – 2,548,714 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.08 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -24.34 |
| Energy contribution | -22.80 |
| Covariance contribution | -1.54 |
| Combinations/Pair | 1.35 |
| Mean z-score | -4.23 |
| Structure conservation index | 0.63 |
| SVM decision value | 7.04 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
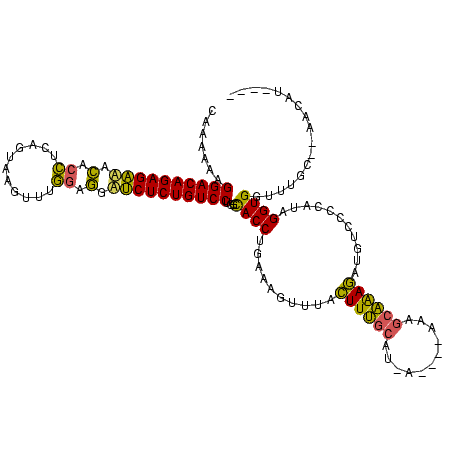
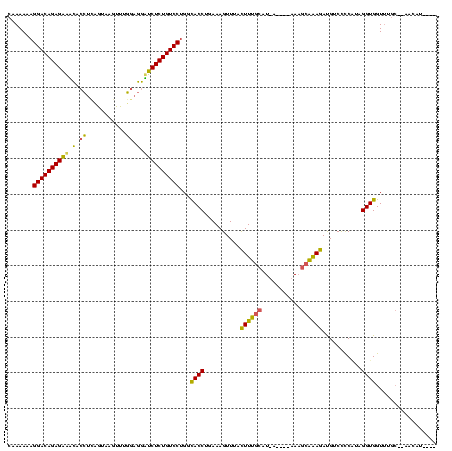
>Bsubt.0 2548606 108 + 4214630/0-120 CAAAAAAGGACAGAGAAACACCUCAUGUAAAAUGAAGGUUCUCUGUCCUGGCACCUGAAAGUUUACUUUGCAU-------ACGCAAAGACGUCCCCAAAGGUGGUUUGCGCAUCC----- ((((..((((((((((...(((((((.....))).))))))))))))))..(((((.........((((((..-------..))))))..........))))).)))).......----- ( -36.01) >Blich.0 2546316 116 + 4222334/0-120 CAAAAAAGGACAGAGAAGCCCCUUUGUCAGUUUGGAGGCUCUCUGUCCUGGCACCUGAAAGUUUACUUUGCAUCA----AAAGCAAAGAUGUCCCCUUUGGUGGUUUGCAAAACAUAAUA ((((..((((((((((.(((((...........)).)))))))))))))..((((.((..(..((((((((....----...)))))).))..)..)).)))).))))............ ( -37.10) >Bhalo.0 2941379 111 + 4202352/0-120 CAAAAAAGGACAGAGCUUUACA-CAGUAAGUUGUGUAAAGCUCUGUCCGGUUACCAGAAAGUUUCUUUCCCCUUAUCGUAAAAAGGAAAUGUCUUCGUAGGUGGUCU----AGUGU---- .......(((((((((((((((-(((....))))))))))))))))))(..((((.(((((...))))).((((........)))).............))))..).----.....---- ( -42.70) >consensus CAAAAAAGGACAGAGAAACACCUCAGUAAGUUUGGAGGAUCUCUGUCCUGGCACCUGAAAGUUUACUUUGCAU_A____AAAGCAAAGAUGUCCCCAUAGGUGGUUUGC__AACAU____ .......((((((((((.(.((...........)).).))))))))))...((((..........((((((...........))))))...........))))................. (-24.34 = -22.80 + -1.54)

| Location | 2,548,606 – 2,548,714 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 61.08 |
| Mean single sequence MFE | -39.60 |
| Consensus MFE | -32.86 |
| Energy contribution | -29.63 |
| Covariance contribution | -3.23 |
| Combinations/Pair | 1.48 |
| Mean z-score | -4.51 |
| Structure conservation index | 0.83 |
| SVM decision value | 6.16 |
| SVM RNA-class probability | 0.999997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
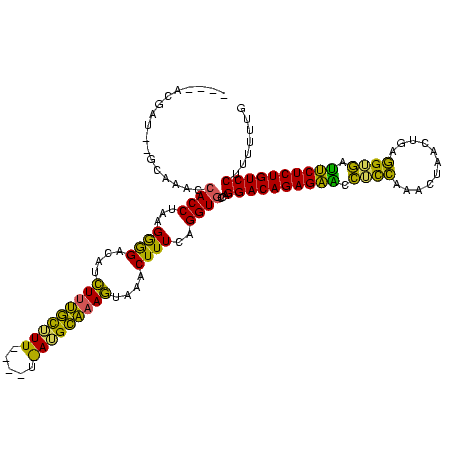
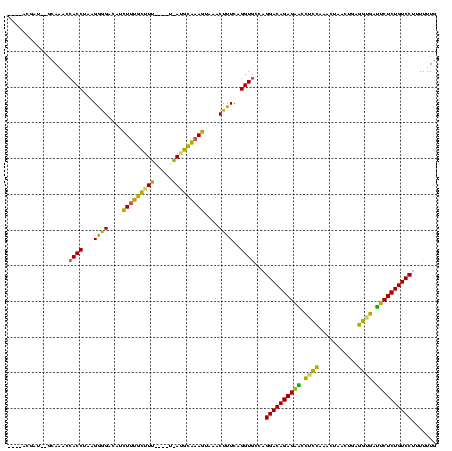
>Bsubt.0 2548606 108 + 4214630/0-120 -----GGAUGCGCAAACCACCUUUGGGGACGUCUUUGCGU-------AUGCAAAGUAAACUUUCAGGUGCCAGGACAGAGAACCUUCAUUUUACAUGAGGUGUUUCUCUGUCCUUUUUUG -----............(((((..(((.....((((((..-------..))))))....)))..)))))..((((((((((((((((((.....)))))).).)))))))))))...... ( -37.50) >Blich.0 2546316 116 + 4222334/0-120 UAUUAUGUUUUGCAAACCACCAAAGGGGACAUCUUUGCUUU----UGAUGCAAAGUAAACUUUCAGGUGCCAGGACAGAGAGCCUCCAAACUGACAAAGGGGCUUCUCUGUCCUUUUUUG .................((((..((((.....((((((...----....))))))....))))..))))..(((((((((((((((............))))).))))))))))...... ( -38.20) >Bhalo.0 2941379 111 + 4202352/0-120 ----ACACU----AGACCACCUACGAAGACAUUUCCUUUUUACGAUAAGGGGAAAGAAACUUUCUGGUAACCGGACAGAGCUUUACACAACUUACUG-UGUAAAGCUCUGUCCUUUUUUG ----.....----.....(((...((((...(((((((((......)))))))))....))))..)))....(((((((((((((((((......))-)))))))))))))))....... ( -43.10) >consensus ____ACGAU__GCAAACCACCUAAGGGGACAUCUUUGCUUU____U_AUGCAAAGUAAACUUUCAGGUGCCAGGACAGAGAACCUCCAAACUAACUGAGGUGAUUCUCUGUCCUUUUUUG .................((((...((((....(((((((((.....)))))))))....))))..))))...((((((((((.((((...........)))).))))))))))....... (-32.86 = -29.63 + -3.23)

| Location | 2,548,606 – 2,548,712 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.14 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -24.14 |
| Energy contribution | -22.52 |
| Covariance contribution | -1.62 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 2548606 106 + 4214630/40-160 GAACUCUUGCUCGACGCAACUCUGGAGAGUGUUUGUGC-------GGAUGCGCAAACCACCUUUGGGGACGUCUUUGCGU-------AUGCAAAGUAAACUUUCAGGUGCCAGGACAGAG ...(((((((.....))).(((..(((.((((((((((-------....))))))).))))))..)))..(((((.(((.-------.((.((((....))))))..))).))))))))) ( -37.80) >Blich.0 2546316 116 + 4222334/40-160 GAACUCUUGCUUGACGCAACUCUGGAGAGAGUUUGUAUCAUAUUAUGUUUUGCAAACCACCAAAGGGGACAUCUUUGCUUU----UGAUGCAAAGUAAACUUUCAGGUGCCAGGACAGAG ...(((((((.....)))..(((((.....(((((((.(((...)))...)))))))((((..((((.....((((((...----....))))))....))))..)))))))))..)))) ( -30.20) >Bhalo.0 2941379 107 + 4202352/40-160 AGACUCUUGCUCGACGCAGCUCUGGAGAGCGUCUA---------ACACU----AGACCACCUACGAAGACAUUUCCUUUUUACGAUAAGGGGAAAGAAACUUUCUGGUAACCGGACAGAG ...((((.(..((.....((((....))))(((((---------....)----)))).(((...((((...(((((((((......)))))))))....))))..)))...))..))))) ( -28.30) >consensus GAACUCUUGCUCGACGCAACUCUGGAGAGAGUUUGU________ACGAU__GCAAACCACCUAAGGGGACAUCUUUGCUUU____U_AUGCAAAGUAAACUUUCAGGUGCCAGGACAGAG ......((((.....))))(((((......(((((((.............)))))))((((...((((....(((((((((.....)))))))))....))))..))))......))))) (-24.14 = -22.52 + -1.62)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:17:27 2006