| Sequence ID | Banth.0 |
|---|---|
| Location | 87,091 – 87,209 |
| Length | 118 |
| Max. P | 0.916980 |

| Location | 87,091 – 87,209 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 88.36 |
| Mean single sequence MFE | -46.04 |
| Consensus MFE | -44.87 |
| Energy contribution | -41.48 |
| Covariance contribution | -3.38 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 87091 118 + 5227293 GUUUGGUGAUGAUGGCAGAGAGGUCACACCCGUUCCCAUACCGAACACGGAAGUUAAGCUCUCUAGCGCCGAUGGUAGUUGGGACCUUGUCCCUGUGAGAGUAGGACGUCGCCAAGCA (((((((((((((((..(((.((......)).))))))).((....((....))...((((((.((((((...))).)))(((((...)))))...)))))).)).))))))))))). ( -42.70) >Bcere.0 87092 118 + 5224283 GUUUGGUGAUGAUGGCAGAGAGGUCACACCCGUUCCCAUACCGAACACGGAAGUUAAGCUCUCUAGCGCCGAUGGUAGUUGGGACCUUGUCCCUGUGAGAGUAGGACGUCGCCAAGCA (((((((((((((((..(((.((......)).))))))).((....((....))...((((((.((((((...))).)))(((((...)))))...)))))).)).))))))))))). ( -42.70) >Bsubt.0 95232 118 + 4214630 GUUUGGUGGCGAUAGCGAAGAGGUCACACCCGUUCCCAUACCGAACACGGAAGUUAAGCUCUUCAGCGCCGAUGGUAGUCGGGGGUUUCCCCCUGUGAGAGUAGGACGCCGCCAAGCA (((((((((((...((((((((.........((((.......))))((....))....)))))).)).(((((....)))(((((...)))))..........)).))))))))))). ( -47.90) >Blich.0 100126 118 + 4222334 GUUUGGUGGCGAUAGCGAAGAGGUCACACCCGUUCCCAUGCCGAACACGGAAGUUAAGCUCUUCAGCGCCGAUGGUAGUUGGGGGCUUCCCCCUGUGAGAGUAGGACGCCGCCAAGCG (((((((((((...((((((((.........((((.......))))((....))....)))))).)).((..........(((((...)))))..........)).))))))))))). ( -46.65) >Bhalo.0 662065 118 + 4202352 GUUUGGUGGCGAUAGCGAAGAGGCCACACCCGUUCCCAUGCCGAACACGGUCGUUAAGCUCUUCAGCGCCGAUGGUAGUUGGGGACUUCCCCCUGCGAGAGUAGGACGCUGCCAAGCA ((((((..(((.....(((..((......))((((((((((((....(((.((((((....)).))))))).)))))..))))))))))..(((((....))))).)))..)))))). ( -48.30) >Bclau.0 57446 118 + 4303871 GUCCGGUGACAAUGGCAAAGAGGUCACACCCGUUCCCAUGCCGAACACGGCCGUUAAGCUCUUUUGCGCCGAUGGUAGUUGGAGGCUUCCUCCUGCGAGAGUAGGACGUUGCCGGGCA ((((((..((.((((....(.((.....)))....))))(((...((..((((((..((......))...))))))...))..)))....((((((....)))))).))..)))))). ( -48.00) >consensus GUUUGGUGACGAUAGCAAAGAGGUCACACCCGUUCCCAUACCGAACACGGAAGUUAAGCUCUUCAGCGCCGAUGGUAGUUGGGGGCUUCCCCCUGUGAGAGUAGGACGCCGCCAAGCA (((((((((((...((((((((.........((((.......))))((....))....)))))).)).((.((....(..(((((...)))))..)....)).)).))))))))))). (-44.87 = -41.48 + -3.38)

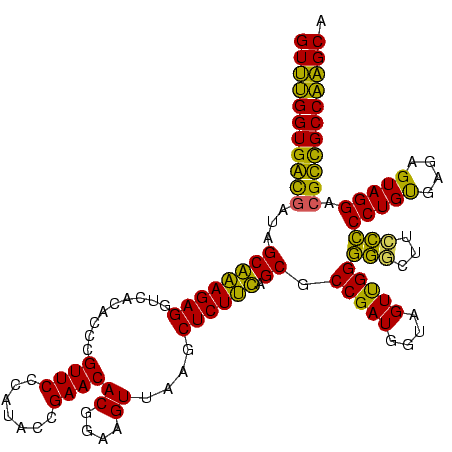
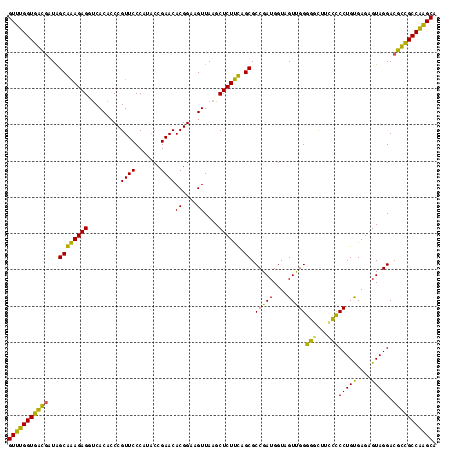
| Location | 87,091 – 87,209 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 88.36 |
| Mean single sequence MFE | -46.04 |
| Consensus MFE | -44.87 |
| Energy contribution | -41.48 |
| Covariance contribution | -3.38 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 87091 118 + 5227293/0-118 GUUUGGUGAUGAUGGCAGAGAGGUCACACCCGUUCCCAUACCGAACACGGAAGUUAAGCUCUCUAGCGCCGAUGGUAGUUGGGACCUUGUCCCUGUGAGAGUAGGACGUCGCCAAGCA (((((((((((((((..(((.((......)).))))))).((....((....))...((((((.((((((...))).)))(((((...)))))...)))))).)).))))))))))). ( -42.70) >Bcere.0 87092 118 + 5224283/0-118 GUUUGGUGAUGAUGGCAGAGAGGUCACACCCGUUCCCAUACCGAACACGGAAGUUAAGCUCUCUAGCGCCGAUGGUAGUUGGGACCUUGUCCCUGUGAGAGUAGGACGUCGCCAAGCA (((((((((((((((..(((.((......)).))))))).((....((....))...((((((.((((((...))).)))(((((...)))))...)))))).)).))))))))))). ( -42.70) >Bsubt.0 95232 118 + 4214630/0-118 GUUUGGUGGCGAUAGCGAAGAGGUCACACCCGUUCCCAUACCGAACACGGAAGUUAAGCUCUUCAGCGCCGAUGGUAGUCGGGGGUUUCCCCCUGUGAGAGUAGGACGCCGCCAAGCA (((((((((((...((((((((.........((((.......))))((....))....)))))).)).(((((....)))(((((...)))))..........)).))))))))))). ( -47.90) >Blich.0 100126 118 + 4222334/0-118 GUUUGGUGGCGAUAGCGAAGAGGUCACACCCGUUCCCAUGCCGAACACGGAAGUUAAGCUCUUCAGCGCCGAUGGUAGUUGGGGGCUUCCCCCUGUGAGAGUAGGACGCCGCCAAGCG (((((((((((...((((((((.........((((.......))))((....))....)))))).)).((..........(((((...)))))..........)).))))))))))). ( -46.65) >Bhalo.0 662065 118 + 4202352/0-118 GUUUGGUGGCGAUAGCGAAGAGGCCACACCCGUUCCCAUGCCGAACACGGUCGUUAAGCUCUUCAGCGCCGAUGGUAGUUGGGGACUUCCCCCUGCGAGAGUAGGACGCUGCCAAGCA ((((((..(((.....(((..((......))((((((((((((....(((.((((((....)).))))))).)))))..))))))))))..(((((....))))).)))..)))))). ( -48.30) >Bclau.0 57446 118 + 4303871/0-118 GUCCGGUGACAAUGGCAAAGAGGUCACACCCGUUCCCAUGCCGAACACGGCCGUUAAGCUCUUUUGCGCCGAUGGUAGUUGGAGGCUUCCUCCUGCGAGAGUAGGACGUUGCCGGGCA ((((((..((.((((....(.((.....)))....))))(((...((..((((((..((......))...))))))...))..)))....((((((....)))))).))..)))))). ( -48.00) >consensus GUUUGGUGACGAUAGCAAAGAGGUCACACCCGUUCCCAUACCGAACACGGAAGUUAAGCUCUUCAGCGCCGAUGGUAGUUGGGGGCUUCCCCCUGUGAGAGUAGGACGCCGCCAAGCA (((((((((((...((((((((.........((((.......))))((....))....)))))).)).((.((....(..(((((...)))))..)....)).)).))))))))))). (-44.87 = -41.48 + -3.38)

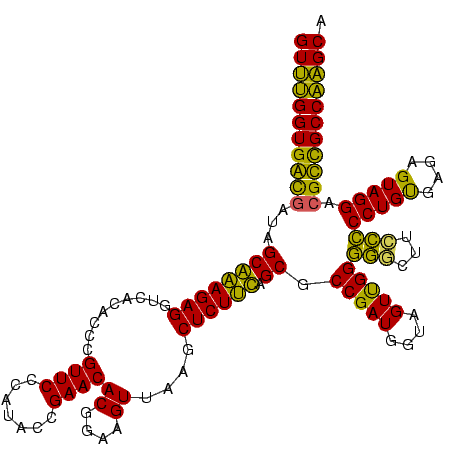
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:17:17 2006