
| Sequence ID | Bcere.0 |
|---|---|
| Location | 3,064,250 – 3,064,332 |
| Length | 82 |
| Max. P | 0.999970 |

| Location | 3,064,250 – 3,064,332 |
|---|---|
| Length | 82 |
| Sequences | 4 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 75.20 |
| Mean single sequence MFE | -17.71 |
| Consensus MFE | -13.31 |
| Energy contribution | -15.31 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.75 |
| SVM decision value | 5.04 |
| SVM RNA-class probability | 0.999970 |
| Prediction | RNA |
| WARNING | Sequence 1: Base composition out of range. |
| WARNING | Sequence 2: Base composition out of range. |
| WARNING | Sequence 4: Base composition out of range. |
Download alignment: ClustalW | MAF
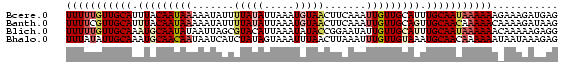
>Bcere.0 3064250 82 + 5224283 UUUUUGUUGCAUUUACAAUAAAAAUAUUUUAUAUUAAAUGUAACUUCAAAUUGUUGCAUUUGCAAUAAAAAAGAAAGAUGAG ((((((((((.......(((((.....)))))...(((((((((........)))))))))))))))))))........... ( -17.60) >Banth.0 3002580 82 + 5227293 UUUUCGUUGCAUUUACAAUAAAAAUAUUUUAUAUUAAAUGUAACUUCAAAUUGUUGCAGUUGCAACAAAAACAAAAGAUAAG .....((((((((((..(((((.....)))))..))))))))))......(((((((....))))))).............. ( -18.60) >Blich.0 3642020 82 - 4222334 UUUUUGUUGCAAAUGCAAUAUAAUUAGCGUACAUUAAAUAUACCGGAAUAUUGUUGCAUUUGCAAUAAAAAACAAAAAGAGG ((((((((((((((((((((..(((...(((.........)))...)))..))))))))))))))))))))........... ( -23.50) >Bhalo.0 3197428 82 + 4202352 UUUAUAUUGCAAAUGCAACAAUAAUCAUCUAUAGUAAAUUUAACUUAAAUUUGUUGUAAAUGCAACAAAAAAUAAUAAAGAG ......(((((..((((((((.............................))))))))..)))))................. ( -11.15) >consensus UUUUUGUUGCAAAUACAAUAAAAAUAUUUUAUAUUAAAUGUAACUUAAAAUUGUUGCAUUUGCAACAAAAAAAAAAAAUAAG (((((((((((.(((((((((.......(((((.....))))).......))))))))).)))))))))))........... (-13.31 = -15.31 + 2.00)


| Location | 3,064,250 – 3,064,332 |
|---|---|
| Length | 82 |
| Sequences | 4 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 75.20 |
| Mean single sequence MFE | -14.27 |
| Consensus MFE | -9.62 |
| Energy contribution | -9.75 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985168 |
| Prediction | RNA |
| WARNING | Sequence 1: Base composition out of range. |
| WARNING | Sequence 2: Base composition out of range. |
| WARNING | Sequence 4: Base composition out of range. |
Download alignment: ClustalW | MAF
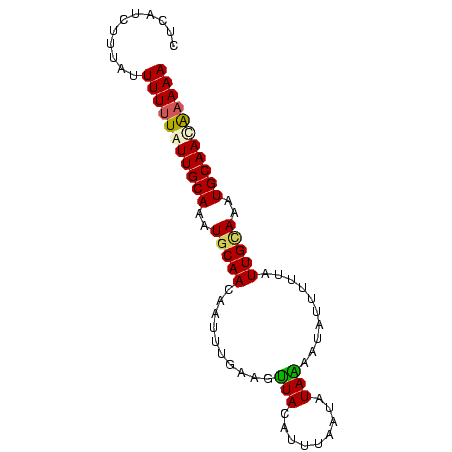
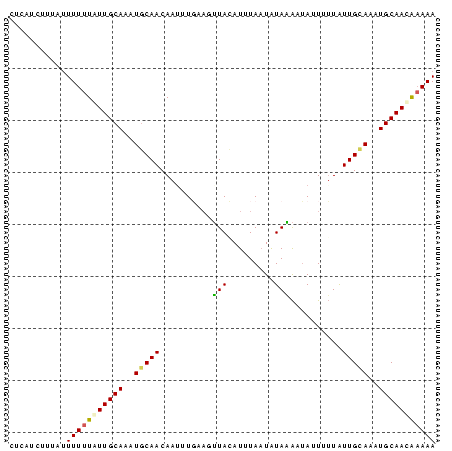
>Bcere.0 3064250 82 + 5224283 CUCAUCUUUCUUUUUUAUUGCAAAUGCAACAAUUUGAAGUUACAUUUAAUAUAAAAUAUUUUUAUUGUAAAUGCAACAAAAA .......(((....((.((((....)))).))...)))(((.((((((..(((((.....)))))..)))))).)))..... ( -10.20) >Banth.0 3002580 82 + 5227293 CUUAUCUUUUGUUUUUGUUGCAACUGCAACAAUUUGAAGUUACAUUUAAUAUAAAAUAUUUUUAUUGUAAAUGCAACGAAAA ......(((((((.(((((((....)))))))..........((((((..(((((.....)))))..)))))).))))))). ( -15.00) >Blich.0 3642020 82 - 4222334 CCUCUUUUUGUUUUUUAUUGCAAAUGCAACAAUAUUCCGGUAUAUUUAAUGUACGCUAAUUAUAUUGCAUUUGCAACAAAAA ...........(((((.((((((((((((....(((...((((((...))))))...)))....)))))))))))).))))) ( -18.60) >Bhalo.0 3197428 82 + 4202352 CUCUUUAUUAUUUUUUGUUGCAUUUACAACAAAUUUAAGUUAAAUUUACUAUAGAUGAUUAUUGUUGCAUUUGCAAUAUAAA ...............(((((((..(.((((((((((((((.......))).))))).....)))))).)..))))))).... ( -13.30) >consensus CUCAUCUUUAUUUUUUAUUGCAAAUGCAACAAUUUGAAGUUACAUUUAAUAUAAAAUAUUUUUAUUGCAAAUGCAACAAAAA ...........(((((((((((..(((((..........(((.........)))..........)))))..))))))))))) ( -9.62 = -9.75 + 0.12)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:17:05 2006