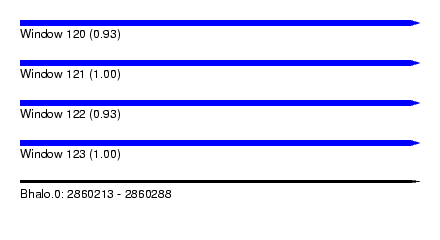
| Sequence ID | Bhalo.0 |
|---|---|
| Location | 2,860,213 – 2,860,288 |
| Length | 75 |
| Max. P | 0.999694 |

| Location | 2,860,213 – 2,860,288 |
|---|---|
| Length | 75 |
| Sequences | 6 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 85.96 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -23.18 |
| Energy contribution | -20.97 |
| Covariance contribution | -2.22 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
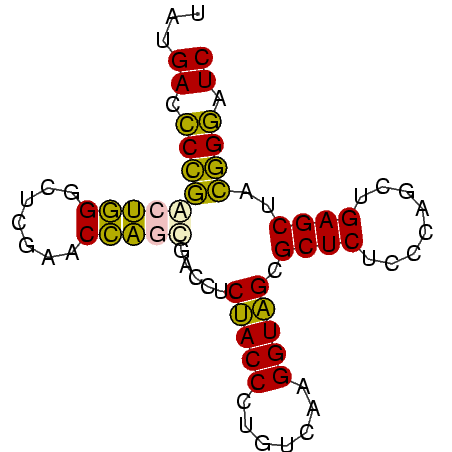
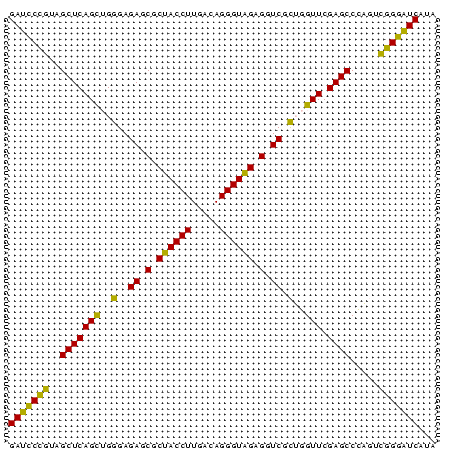
>Bhalo.0 2860213 75 + 4202352 AAUGACCCGUACUGGGCUCGAACCAGUGACCUCUACCCUGUCAAGGUAGCGCUCUCCCAACUGAGCUAACGGAUC ......(((((((((.......))))).....(((((.......))))).((((........))))..))))... ( -21.70) >Bclau.0 2571144 75 + 4303871 UAUGACCCGUACUGGGCUCGAACCAGUGACCUCUACCCUGUCAAGGUAGCGCUCUCCCAACUGAGCUAACGGAUC ......(((((((((.......))))).....(((((.......))))).((((........))))..))))... ( -21.70) >Bsubt.0 2952455 75 - 4214630 UAUGAUUCCGACUGGGCUCGAACCAGCGACCUCUACCCUGUCAAGGUAGCGCUCUCCCAGCUGAGCUACGGAAUC ...(((((((....((((((.....((.((((..((...))..)))).))(((.....))))))))).))))))) ( -24.60) >Blich.0 1299724 75 + 4222334 UAUGAUUCCGACUGGGCUCGAACCAGCGACCUCUACCCUGUCAAGGUAGCGCUCUCCCAGCUGAGCUACGGAAUC ...(((((((....((((((.....((.((((..((...))..)))).))(((.....))))))))).))))))) ( -24.60) >Bcere.0 1389507 75 + 5224283 UAUGACCCCGGAGAGGCUCGAACUCACGACCUCCACCCUGUCAAGGUGGCGCUCUCCCUGCUGAGCUACGGGAUC ......((((..((((.(((......)))))))((((.......))))..((((........))))..))))... ( -25.00) >Banth.0 1232475 75 + 5227293 UAUGAUCCCGGAGAGGCUCGAACUCACGACCUCCACCCUGUCAAGGUGGCGCUCUCCCUGCUGAGCUACGGGAUC ...(((((((..((((.(((......)))))))((((.......))))..((((........))))..))))))) ( -29.50) >consensus UAUGACCCCGACUGGGCUCGAACCAGCGACCUCUACCCUGUCAAGGUAGCGCUCUCCCAGCUGAGCUACGGGAUC ...((.(((((((((.......))))).....(((((.......))))).((((........))))..)))).)) (-23.18 = -20.97 + -2.22)

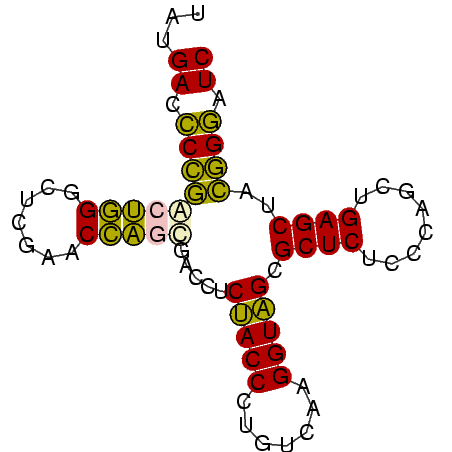
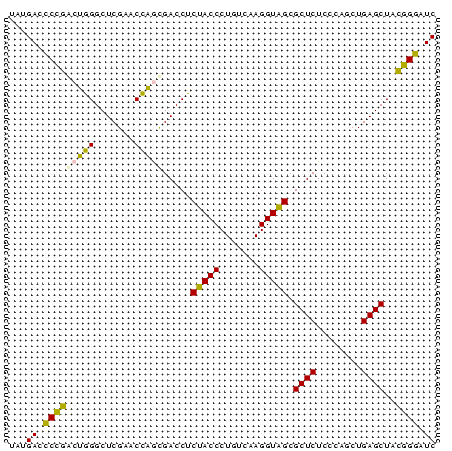
| Location | 2,860,213 – 2,860,288 |
|---|---|
| Length | 75 |
| Sequences | 6 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 85.96 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -33.37 |
| Energy contribution | -30.90 |
| Covariance contribution | -2.47 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.70 |
| Structure conservation index | 1.04 |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
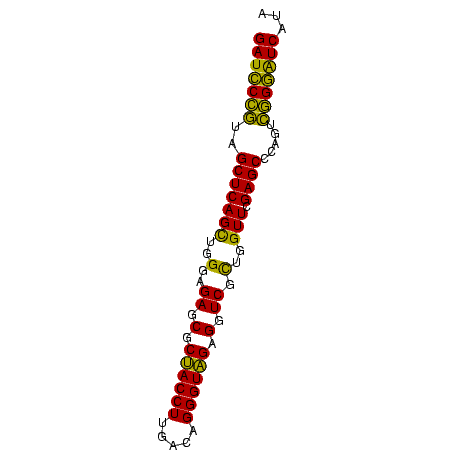
>Bhalo.0 2860213 75 + 4202352 GAUCCGUUAGCUCAGUUGGGAGAGCGCUACCUUGACAGGGUAGAGGUCACUGGUUCGAGCCCAGUACGGGUCAUU (((((((..((((((..((..((.(.((((((.....)))))).).)).))..)).)))).....)))))))... ( -30.00) >Bclau.0 2571144 75 + 4303871 GAUCCGUUAGCUCAGUUGGGAGAGCGCUACCUUGACAGGGUAGAGGUCACUGGUUCGAGCCCAGUACGGGUCAUA (((((((..((((((..((..((.(.((((((.....)))))).).)).))..)).)))).....)))))))... ( -30.00) >Bsubt.0 2952455 75 - 4214630 GAUUCCGUAGCUCAGCUGGGAGAGCGCUACCUUGACAGGGUAGAGGUCGCUGGUUCGAGCCCAGUCGGAAUCAUA (((((((..(((((((..(..((.(.((((((.....)))))).).)).)..))).)))).....)))))))... ( -33.40) >Blich.0 1299724 75 + 4222334 GAUUCCGUAGCUCAGCUGGGAGAGCGCUACCUUGACAGGGUAGAGGUCGCUGGUUCGAGCCCAGUCGGAAUCAUA (((((((..(((((((..(..((.(.((((((.....)))))).).)).)..))).)))).....)))))))... ( -33.40) >Bcere.0 1389507 75 + 5224283 GAUCCCGUAGCUCAGCAGGGAGAGCGCCACCUUGACAGGGUGGAGGUCGUGAGUUCGAGCCUCUCCGGGGUCAUA ((((((...((...))..((((((..((((((.....)))))).(.(((......))).)))))))))))))... ( -32.90) >Banth.0 1232475 75 + 5227293 GAUCCCGUAGCUCAGCAGGGAGAGCGCCACCUUGACAGGGUGGAGGUCGUGAGUUCGAGCCUCUCCGGGAUCAUA ((((((...((...))..((((((..((((((.....)))))).(.(((......))).)))))))))))))... ( -33.50) >consensus GAUCCCGUAGCUCAGCUGGGAGAGCGCUACCUUGACAGGGUAGAGGUCGCUGGUUCGAGCCCAGUCGGGAUCAUA (((((((..(((((((..(..((.(.((((((.....)))))).).)).)..))).)))).....)))))))... (-33.37 = -30.90 + -2.47)

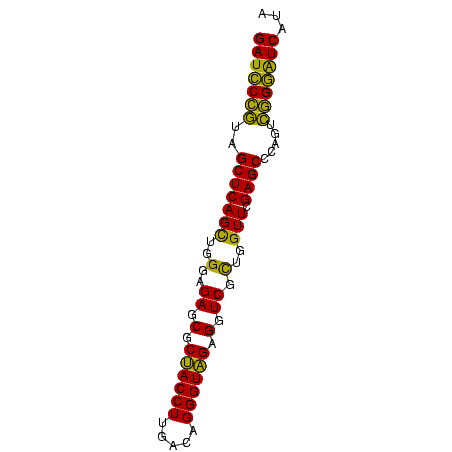
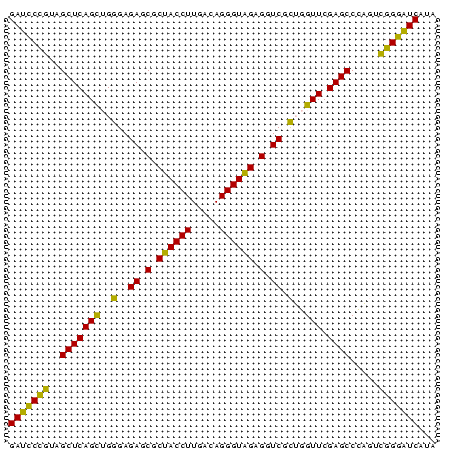
| Location | 2,860,213 – 2,860,288 |
|---|---|
| Length | 75 |
| Sequences | 6 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 85.96 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -23.18 |
| Energy contribution | -20.97 |
| Covariance contribution | -2.22 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 2860213 75 + 4202352/0-75 AAUGACCCGUACUGGGCUCGAACCAGUGACCUCUACCCUGUCAAGGUAGCGCUCUCCCAACUGAGCUAACGGAUC ......(((((((((.......))))).....(((((.......))))).((((........))))..))))... ( -21.70) >Bclau.0 2571144 75 + 4303871/0-75 UAUGACCCGUACUGGGCUCGAACCAGUGACCUCUACCCUGUCAAGGUAGCGCUCUCCCAACUGAGCUAACGGAUC ......(((((((((.......))))).....(((((.......))))).((((........))))..))))... ( -21.70) >Bsubt.0 2952455 75 - 4214630/0-75 UAUGAUUCCGACUGGGCUCGAACCAGCGACCUCUACCCUGUCAAGGUAGCGCUCUCCCAGCUGAGCUACGGAAUC ...(((((((....((((((.....((.((((..((...))..)))).))(((.....))))))))).))))))) ( -24.60) >Blich.0 1299724 75 + 4222334/0-75 UAUGAUUCCGACUGGGCUCGAACCAGCGACCUCUACCCUGUCAAGGUAGCGCUCUCCCAGCUGAGCUACGGAAUC ...(((((((....((((((.....((.((((..((...))..)))).))(((.....))))))))).))))))) ( -24.60) >Bcere.0 1389507 75 + 5224283/0-75 UAUGACCCCGGAGAGGCUCGAACUCACGACCUCCACCCUGUCAAGGUGGCGCUCUCCCUGCUGAGCUACGGGAUC ......((((..((((.(((......)))))))((((.......))))..((((........))))..))))... ( -25.00) >Banth.0 1232475 75 + 5227293/0-75 UAUGAUCCCGGAGAGGCUCGAACUCACGACCUCCACCCUGUCAAGGUGGCGCUCUCCCUGCUGAGCUACGGGAUC ...(((((((..((((.(((......)))))))((((.......))))..((((........))))..))))))) ( -29.50) >consensus UAUGACCCCGACUGGGCUCGAACCAGCGACCUCUACCCUGUCAAGGUAGCGCUCUCCCAGCUGAGCUACGGGAUC ...((.(((((((((.......))))).....(((((.......))))).((((........))))..)))).)) (-23.18 = -20.97 + -2.22)

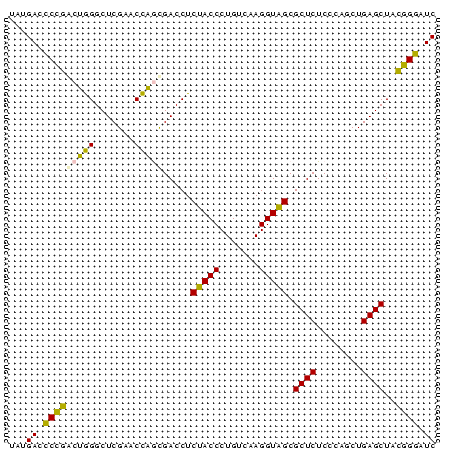
| Location | 2,860,213 – 2,860,288 |
|---|---|
| Length | 75 |
| Sequences | 6 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 85.96 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -33.37 |
| Energy contribution | -30.90 |
| Covariance contribution | -2.47 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.70 |
| Structure conservation index | 1.04 |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 2860213 75 + 4202352/0-75 GAUCCGUUAGCUCAGUUGGGAGAGCGCUACCUUGACAGGGUAGAGGUCACUGGUUCGAGCCCAGUACGGGUCAUU (((((((..((((((..((..((.(.((((((.....)))))).).)).))..)).)))).....)))))))... ( -30.00) >Bclau.0 2571144 75 + 4303871/0-75 GAUCCGUUAGCUCAGUUGGGAGAGCGCUACCUUGACAGGGUAGAGGUCACUGGUUCGAGCCCAGUACGGGUCAUA (((((((..((((((..((..((.(.((((((.....)))))).).)).))..)).)))).....)))))))... ( -30.00) >Bsubt.0 2952455 75 - 4214630/0-75 GAUUCCGUAGCUCAGCUGGGAGAGCGCUACCUUGACAGGGUAGAGGUCGCUGGUUCGAGCCCAGUCGGAAUCAUA (((((((..(((((((..(..((.(.((((((.....)))))).).)).)..))).)))).....)))))))... ( -33.40) >Blich.0 1299724 75 + 4222334/0-75 GAUUCCGUAGCUCAGCUGGGAGAGCGCUACCUUGACAGGGUAGAGGUCGCUGGUUCGAGCCCAGUCGGAAUCAUA (((((((..(((((((..(..((.(.((((((.....)))))).).)).)..))).)))).....)))))))... ( -33.40) >Bcere.0 1389507 75 + 5224283/0-75 GAUCCCGUAGCUCAGCAGGGAGAGCGCCACCUUGACAGGGUGGAGGUCGUGAGUUCGAGCCUCUCCGGGGUCAUA ((((((...((...))..((((((..((((((.....)))))).(.(((......))).)))))))))))))... ( -32.90) >Banth.0 1232475 75 + 5227293/0-75 GAUCCCGUAGCUCAGCAGGGAGAGCGCCACCUUGACAGGGUGGAGGUCGUGAGUUCGAGCCUCUCCGGGAUCAUA ((((((...((...))..((((((..((((((.....)))))).(.(((......))).)))))))))))))... ( -33.50) >consensus GAUCCCGUAGCUCAGCUGGGAGAGCGCUACCUUGACAGGGUAGAGGUCGCUGGUUCGAGCCCAGUCGGGAUCAUA (((((((..(((((((..(..((.(.((((((.....)))))).).)).)..))).)))).....)))))))... (-33.37 = -30.90 + -2.47)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:17:00 2006