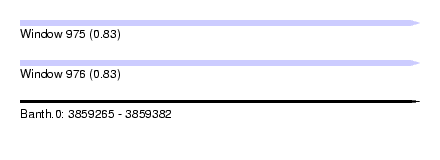
| Sequence ID | Banth.0 |
|---|---|
| Location | 3,859,265 – 3,859,382 |
| Length | 117 |
| Max. P | 0.834368 |

| Location | 3,859,265 – 3,859,382 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.57 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -22.75 |
| Energy contribution | -19.88 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 3859265 117 + 5227293/0-120 UUUAAAAGGGAAGCUUGGUGAAACUCCAACACGGUCCCGCCACUGUAAAUGCUGAGAUUUCUUUUUGAUA-CCACUGUGAAA--ACGGGAAGGUAAAAGAAAUUAUAUGAAGCAUAAGUC .......((((...((((.......)))).....))))...(((....(((((..(((((((((....((-((.((((....--))))...)))))))))))))......))))).))). ( -28.00) >Bcere.0 3791566 117 + 5224283/0-120 UUUAAAAGGGAAGCUUGGUGAAACUCCAACACGGUCCCGCCACUGUAAAUGCUGAGAUUUCUUUUUAAUG-CCACUGUGAAA--ACGGGAAGGCGAAAGAAAUCAUAUGAAGCAUAAGUC .......((((...((((.......)))).....))))...(((....(((((..((((((((((....(-((.((((....--))))...)))))))))))))......))))).))). ( -31.50) >Bhalo.0 1661087 116 + 4202352/0-120 CGUGAAAAGGGAAGUCGGUGAAAAUCCGACACGGUCCCGCCACUGUAAAUGG-GAGAGGCUUGCAAGAU--CCACUGUCUAGCGACGGGAAGGGGGCAAGUACUCGAUGAA-CAUAAGUC .(((....((((.((((((....).)))))....))))..))).....(((.-..(((((((((....(--((.(((((....)))))...))).)))))).)))......-)))..... ( -36.50) >Blich.0 2804747 119 - 4222334/0-120 UUUAAAAGGGAAGUUUGGUGAAAAUCCAACGCGGUCCCGCCACUGUGAAUGAGGAGGUUAUUUCAUAAAACCCACUGUUUCUAUAUGGGAAGGGGGAAAUAACCGU-CGAUUCAUGAGCC .......((((.(((.(((....))).)))....))))((...(((((((..(..(((((((((......((((.(((....)))))))......)))))))))..-).))))))).)). ( -34.70) >consensus UUUAAAAGGGAAGCUUGGUGAAAAUCCAACACGGUCCCGCCACUGUAAAUGCUGAGAUUUCUUCAUAAUA_CCACUGUGAAA__ACGGGAAGGGGAAAAAAACCAUAUGAAGCAUAAGUC .......((((...((((.......)))).....))))..........(((((..(((((((((.......((.((((......))))...))..)))))))))......)))))..... (-22.75 = -19.88 + -2.88)

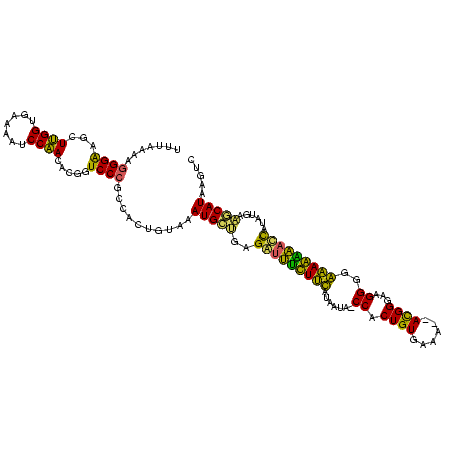
| Location | 3,859,265 – 3,859,382 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.08 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -26.96 |
| Energy contribution | -24.27 |
| Covariance contribution | -2.69 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 3859265 117 + 5227293/13-133 CUUGGUGAAACUCCAACACGGUCCCGCCACUGUAAAUGCUGAGAUUUCUUUUUGAUA-CCACUGUGAAA--ACGGGAAGGUAAAAGAAAUUAUAUGAAGCAUAAGUCAGGAGACCUGCCU .((((.......))))...((((((...(((....(((((..(((((((((....((-((.((((....--))))...)))))))))))))......))))).)))..)).))))..... ( -31.20) >Bcere.0 3791566 117 + 5224283/13-133 CUUGGUGAAACUCCAACACGGUCCCGCCACUGUAAAUGCUGAGAUUUCUUUUUAAUG-CCACUGUGAAA--ACGGGAAGGCGAAAGAAAUCAUAUGAAGCAUAAGUCAGGAGACCUGCCU .((((.......))))...((((((...(((....(((((..((((((((((....(-((.((((....--))))...)))))))))))))......))))).)))..)).))))..... ( -34.70) >Bhalo.0 1661087 116 + 4202352/13-133 GUCGGUGAAAAUCCGACACGGUCCCGCCACUGUAAAUGG-GAGAGGCUUGCAAGAU--CCACUGUCUAGCGACGGGAAGGGGGCAAGUACUCGAUGAA-CAUAAGUCAGGAGACCUGCCU ((((((....).)))))..((((((.(((.......)))-..(((((((((....(--((.(((((....)))))...))).)))))).)))(((...-.....))).)).))))..... ( -39.30) >Blich.0 2804747 119 - 4222334/13-133 UUUGGUGAAAAUCCAACGCGGUCCCGCCACUGUGAAUGAGGAGGUUAUUUCAUAAAACCCACUGUUUCUAUAUGGGAAGGGGGAAAUAACCGU-CGAUUCAUGAGCCAGGAGACCUGCCU .(((((....).)))).((((((((....((((((((..(..(((((((((......((((.(((....)))))))......)))))))))..-).)))))).))...)).)))).)).. ( -36.30) >consensus CUUGGUGAAAAUCCAACACGGUCCCGCCACUGUAAAUGCUGAGAUUUCUUCAUAAUA_CCACUGUGAAA__ACGGGAAGGGGAAAAAAACCAUAUGAAGCAUAAGUCAGGAGACCUGCCU .((((.......))))...((((((...(((....(((((..(((((((((.......((.((((......))))...))..)))))))))......))))).)))..)).))))..... (-26.96 = -24.27 + -2.69)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:13:32 2006