
| Sequence ID | Banth.0 |
|---|---|
| Location | 3,795,570 – 3,795,665 |
| Length | 95 |
| Max. P | 0.992512 |

| Location | 3,795,570 – 3,795,665 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 85.19 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -4.00 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
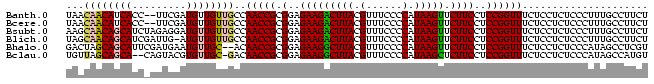
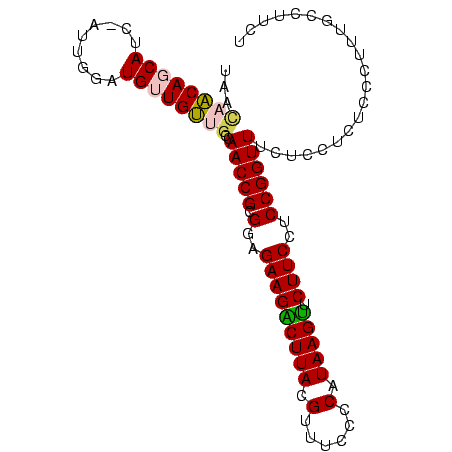
>Banth.0 3795570 95 + 5227293 UAACAACAUCACC--UUCGAUGUUGUUGCCAACCGCGGAGAAGACUUACGUUUCCCCAUAAGUUCUUCCUCCGGUUUCUCCUCUCCCUUUGCCUUCU ((((((((((...--...))))))))))..(((((.(..(((((((((.(......).)))).)))))..))))))..................... ( -25.70) >Bcere.0 3729175 95 + 5224283 UAACAACAUCACC--UUCGAUGUUGUUGCCAACCGCGGAGAAGACUUACGUUUCCCCAUAAGUUCUUCCUCCGGUUUCUCCUCUCCCUUUGCCUUCU ((((((((((...--...))))))))))..(((((.(..(((((((((.(......).)))).)))))..))))))..................... ( -25.70) >Bsubt.0 2645968 97 - 4214630 AAGCAACAGCAUCUAGAGGAUGUUGUUGCCAACCGCGGAGAAGACUUACGUUUCCCCAUAAGUUCUUCCUCCGGUUUCUCCUCUCCCUUUGCCUUCU ..(((((((((((.....))))))))))).(((((.(..(((((((((.(......).)))).)))))..))))))..................... ( -31.30) >Blich.0 2549428 96 - 4222334 UAGCAACAGCAUCGAUUG-AUGUUGUUGCCAACCGCGGAGAAGACUUACGUUUCCCCAUAAGUUCUUCCUCCGGUUUCUCCUCUCCCUUUGCCUUCU ..(((((((((((....)-)))))))))).(((((.(..(((((((((.(......).)))).)))))..))))))..................... ( -30.60) >Bhalo.0 2714057 95 + 4202352 GACUAGCAGCAUUCGAUGAAUGUUGC--ACAACCGCGGAGAAGGCUUACGUUUCCCCAUAAGUUCUUCCUCCGGUUUCUCCUCUCCCAUAGCCUCGU ..((((((((((((...)))))))))--..(((((.(..(((((((((.(......).)))).)))))..))))))............)))...... ( -25.70) >Bclau.0 2508550 94 + 4303871 UGUUAGCAGCA--CAGUACGUGUUGC-GACAACCGCGGAGAAGGCUUACGUUUCCCCAUAAGCUCUUCCUCCGGUUUCUCCUCUCCCAUAGCCAUGU .((((((((((--(.....)))))))-((.(((((.(..(((((((((.(......).))))).))))..))))))))..........))))..... ( -29.80) >consensus UAACAACAGCAUC_AUUGGAUGUUGUUGCCAACCGCGGAGAAGACUUACGUUUCCCCAUAAGUUCUUCCUCCGGUUUCUCCUCUCCCUUUGCCUUCU ...((((((((.........))))))))..(((((.(..(((((((((.(......).))))).))))..))))))..................... (-20.18 = -20.10 + -0.08)
| Location | 3,795,570 – 3,795,665 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 85.19 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -28.90 |
| Energy contribution | -28.52 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
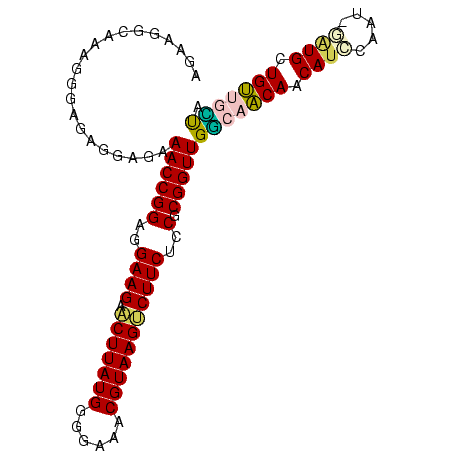
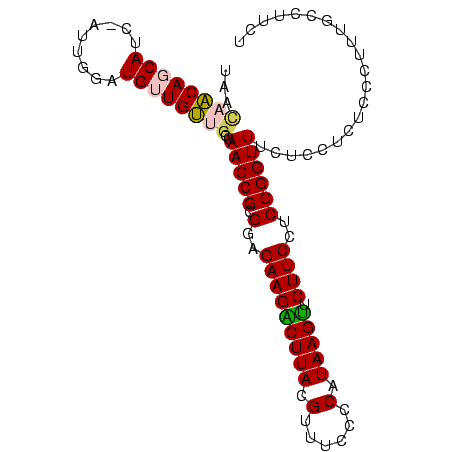
>Banth.0 3795570 95 + 5227293 AGAAGGCAAAGGGAGAGGAGAAACCGGAGGAAGAACUUAUGGGGAAACGUAAGUCUUCUCCGCGGUUGGCAACAACAUCGAA--GGUGAUGUUGUUA ....(.(((.(((((((((....((((((......))).)))(....).....)))))))).)..))).)((((((((((..--..)))))))))). ( -32.10) >Bcere.0 3729175 95 + 5224283 AGAAGGCAAAGGGAGAGGAGAAACCGGAGGAAGAACUUAUGGGGAAACGUAAGUCUUCUCCGCGGUUGGCAACAACAUCGAA--GGUGAUGUUGUUA ....(.(((.(((((((((....((((((......))).)))(....).....)))))))).)..))).)((((((((((..--..)))))))))). ( -32.10) >Bsubt.0 2645968 97 - 4214630 AGAAGGCAAAGGGAGAGGAGAAACCGGAGGAAGAACUUAUGGGGAAACGUAAGUCUUCUCCGCGGUUGGCAACAACAUCCUCUAGAUGCUGUUGCUU ...((((((.((.((((((....((((((.((((.((((((......))))))))))))))).)((((....)))).)))))).....)).)))))) ( -38.20) >Blich.0 2549428 96 - 4222334 AGAAGGCAAAGGGAGAGGAGAAACCGGAGGAAGAACUUAUGGGGAAACGUAAGUCUUCUCCGCGGUUGGCAACAACAU-CAAUCGAUGCUGUUGCUA .....................((((((..(((((.((((((......)))))))))))..).)))))(((((((.(((-(....)))).))))))). ( -34.30) >Bhalo.0 2714057 95 + 4202352 ACGAGGCUAUGGGAGAGGAGAAACCGGAGGAAGAACUUAUGGGGAAACGUAAGCCUUCUCCGCGGUUGU--GCAACAUUCAUCGAAUGCUGCUAGUC ....(((((............((((((..((((..((((((......)))))).))))..).)))))..--(((.(((((...))))).)))))))) ( -27.20) >Bclau.0 2508550 94 + 4303871 ACAUGGCUAUGGGAGAGGAGAAACCGGAGGAAGAGCUUAUGGGGAAACGUAAGCCUUCUCCGCGGUUGUC-GCAACACGUACUG--UGCUGCUAACA .(((((.(((((..(.((...((((((..((((.(((((((......)))))))))))..).))))).))-.)..).)))))))--))......... ( -30.50) >consensus AGAAGGCAAAGGGAGAGGAGAAACCGGAGGAAGAACUUAUGGGGAAACGUAAGUCUUCUCCGCGGUUGGCAACAACAUCCAAU_GAUGCUGUUGCUA .....................((((((..((((.(((((((......)))))))))))..).)))))(((((((.((((.....)))).))))))). (-28.90 = -28.52 + -0.39)

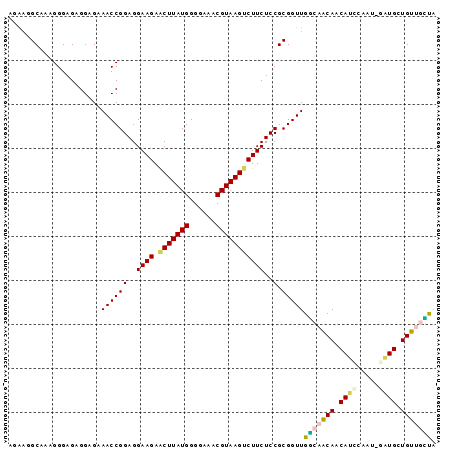
| Location | 3,795,570 – 3,795,665 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 85.19 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -4.00 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 3795570 95 + 5227293/0-97 UAACAACAUCACC--UUCGAUGUUGUUGCCAACCGCGGAGAAGACUUACGUUUCCCCAUAAGUUCUUCCUCCGGUUUCUCCUCUCCCUUUGCCUUCU ((((((((((...--...))))))))))..(((((.(..(((((((((.(......).)))).)))))..))))))..................... ( -25.70) >Bcere.0 3729175 95 + 5224283/0-97 UAACAACAUCACC--UUCGAUGUUGUUGCCAACCGCGGAGAAGACUUACGUUUCCCCAUAAGUUCUUCCUCCGGUUUCUCCUCUCCCUUUGCCUUCU ((((((((((...--...))))))))))..(((((.(..(((((((((.(......).)))).)))))..))))))..................... ( -25.70) >Bsubt.0 2645968 97 - 4214630/0-97 AAGCAACAGCAUCUAGAGGAUGUUGUUGCCAACCGCGGAGAAGACUUACGUUUCCCCAUAAGUUCUUCCUCCGGUUUCUCCUCUCCCUUUGCCUUCU ..(((((((((((.....))))))))))).(((((.(..(((((((((.(......).)))).)))))..))))))..................... ( -31.30) >Blich.0 2549428 96 - 4222334/0-97 UAGCAACAGCAUCGAUUG-AUGUUGUUGCCAACCGCGGAGAAGACUUACGUUUCCCCAUAAGUUCUUCCUCCGGUUUCUCCUCUCCCUUUGCCUUCU ..(((((((((((....)-)))))))))).(((((.(..(((((((((.(......).)))).)))))..))))))..................... ( -30.60) >Bhalo.0 2714057 95 + 4202352/0-97 GACUAGCAGCAUUCGAUGAAUGUUGC--ACAACCGCGGAGAAGGCUUACGUUUCCCCAUAAGUUCUUCCUCCGGUUUCUCCUCUCCCAUAGCCUCGU ..((((((((((((...)))))))))--..(((((.(..(((((((((.(......).)))).)))))..))))))............)))...... ( -25.70) >Bclau.0 2508550 94 + 4303871/0-97 UGUUAGCAGCA--CAGUACGUGUUGC-GACAACCGCGGAGAAGGCUUACGUUUCCCCAUAAGCUCUUCCUCCGGUUUCUCCUCUCCCAUAGCCAUGU .((((((((((--(.....)))))))-((.(((((.(..(((((((((.(......).))))).))))..))))))))..........))))..... ( -29.80) >consensus UAACAACAGCAUC_AUUGGAUGUUGUUGCCAACCGCGGAGAAGACUUACGUUUCCCCAUAAGUUCUUCCUCCGGUUUCUCCUCUCCCUUUGCCUUCU ...((((((((.........))))))))..(((((.(..(((((((((.(......).))))).))))..))))))..................... (-20.18 = -20.10 + -0.08)
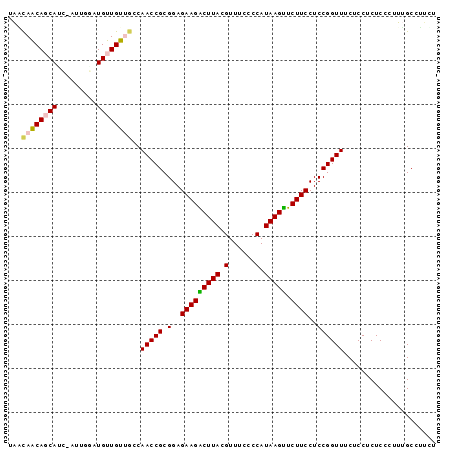
| Location | 3,795,570 – 3,795,665 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 85.19 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -28.90 |
| Energy contribution | -28.52 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 3795570 95 + 5227293/0-97 AGAAGGCAAAGGGAGAGGAGAAACCGGAGGAAGAACUUAUGGGGAAACGUAAGUCUUCUCCGCGGUUGGCAACAACAUCGAA--GGUGAUGUUGUUA ....(.(((.(((((((((....((((((......))).)))(....).....)))))))).)..))).)((((((((((..--..)))))))))). ( -32.10) >Bcere.0 3729175 95 + 5224283/0-97 AGAAGGCAAAGGGAGAGGAGAAACCGGAGGAAGAACUUAUGGGGAAACGUAAGUCUUCUCCGCGGUUGGCAACAACAUCGAA--GGUGAUGUUGUUA ....(.(((.(((((((((....((((((......))).)))(....).....)))))))).)..))).)((((((((((..--..)))))))))). ( -32.10) >Bsubt.0 2645968 97 - 4214630/0-97 AGAAGGCAAAGGGAGAGGAGAAACCGGAGGAAGAACUUAUGGGGAAACGUAAGUCUUCUCCGCGGUUGGCAACAACAUCCUCUAGAUGCUGUUGCUU ...((((((.((.((((((....((((((.((((.((((((......))))))))))))))).)((((....)))).)))))).....)).)))))) ( -38.20) >Blich.0 2549428 96 - 4222334/0-97 AGAAGGCAAAGGGAGAGGAGAAACCGGAGGAAGAACUUAUGGGGAAACGUAAGUCUUCUCCGCGGUUGGCAACAACAU-CAAUCGAUGCUGUUGCUA .....................((((((..(((((.((((((......)))))))))))..).)))))(((((((.(((-(....)))).))))))). ( -34.30) >Bhalo.0 2714057 95 + 4202352/0-97 ACGAGGCUAUGGGAGAGGAGAAACCGGAGGAAGAACUUAUGGGGAAACGUAAGCCUUCUCCGCGGUUGU--GCAACAUUCAUCGAAUGCUGCUAGUC ....(((((............((((((..((((..((((((......)))))).))))..).)))))..--(((.(((((...))))).)))))))) ( -27.20) >Bclau.0 2508550 94 + 4303871/0-97 ACAUGGCUAUGGGAGAGGAGAAACCGGAGGAAGAGCUUAUGGGGAAACGUAAGCCUUCUCCGCGGUUGUC-GCAACACGUACUG--UGCUGCUAACA .(((((.(((((..(.((...((((((..((((.(((((((......)))))))))))..).))))).))-.)..).)))))))--))......... ( -30.50) >consensus AGAAGGCAAAGGGAGAGGAGAAACCGGAGGAAGAACUUAUGGGGAAACGUAAGUCUUCUCCGCGGUUGGCAACAACAUCCAAU_GAUGCUGUUGCUA .....................((((((..((((.(((((((......)))))))))))..).)))))(((((((.((((.....)))).))))))). (-28.90 = -28.52 + -0.39)

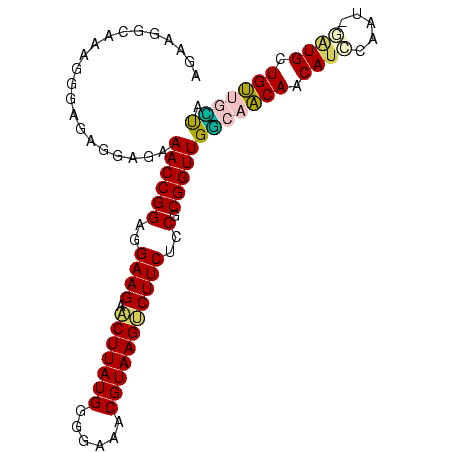
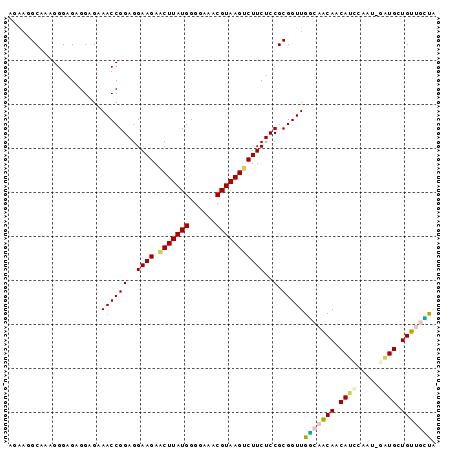
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:28:13 2006