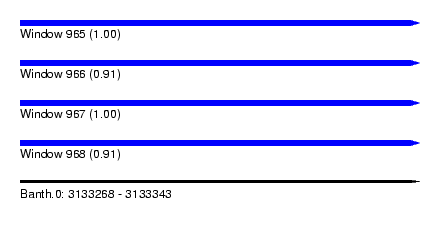
| Sequence ID | Banth.0 |
|---|---|
| Location | 3,133,268 – 3,133,343 |
| Length | 75 |
| Max. P | 0.997183 |

| Location | 3,133,268 – 3,133,343 |
|---|---|
| Length | 75 |
| Sequences | 6 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 68.13 |
| Mean single sequence MFE | -18.40 |
| Consensus MFE | -10.88 |
| Energy contribution | -10.05 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
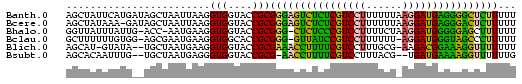
>Banth.0 3133268 75 + 5227293 AAAAAGAGCCCCUCAUCCUUAAAAAAGGACGAGAGACUCCCGCGGUACCACCUUAAUUAGCUAUCAUGAAUAGCU .....(((.(.(((.(((((....))))).))).).)))....((.....))......((((((.....)))))) ( -17.20) >Bcere.0 3157045 74 + 5224283 AAAAAGAGUCCCUCAUCCUUAAAAAAGGACGAGAGACUCCCGCGGUACCACCUUAAUUAGCUAUC-UUUAUAGCU .....(((((.(((.(((((....))))).))).)))))....((.....))......((((((.-...)))))) ( -22.00) >Bhalo.0 2322309 72 + 4202352 AAAAAAGCUCCCCCAUCCUUAGAAAAGGACGGGAGAG-CCCGCGGUACCACCUUCAUU-GGU-CAAUAAAUAACC ......((((.(((.(((((....))))).))).)))-).....(.((((.......)-)))-)........... ( -19.90) >Bclau.0 3999426 72 - 4303871 AAAAAGGGCUACCCAUCCU-AAAAAAGGACGGAUAAC-CCCGCGGUGCCACCUUCAUUCGCU-CCACAAAAAAGC .....(((.((.((.((((-.....)))).)).)).)-)).(.((.((...........)).-)).)........ ( -15.30) >Blich.0 1756018 71 - 4222334 AAAAAAACCUUUCCGUCUU-CGCAAAGGACGAAAAGGUUUCGCGGUACCACCUUCAUUAGCA--UAUAC-AUGCU ....((((((((.((((((-.....)))))).))))))))...((.....))......((((--(....-))))) ( -18.50) >Bsubt.0 1742314 70 - 4214630 CAAAAAACCUUUUCAUCA--CGUAAAGGACGAAAAGGUU-CGCGGUACCACCCUCAUUAGCA--CAAAUUGUGCU .....(((((((((.((.--(.....))).)))))))))-.(.((......)).)...((((--(.....))))) ( -17.50) >consensus AAAAAAACCCCCCCAUCCU_AAAAAAGGACGAAAAACUCCCGCGGUACCACCUUCAUUAGCU__CAUAAAUAGCU .....(((((.(((.((((......)))).))).)))))....((.....))....................... (-10.88 = -10.05 + -0.83)

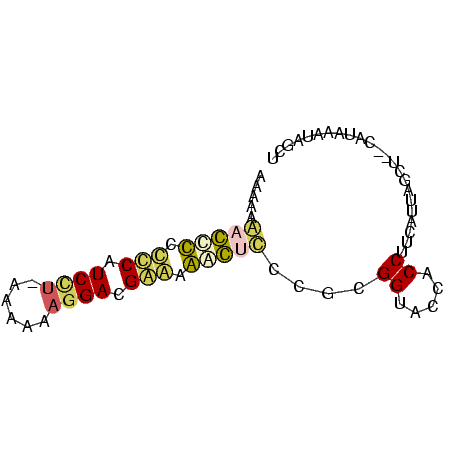
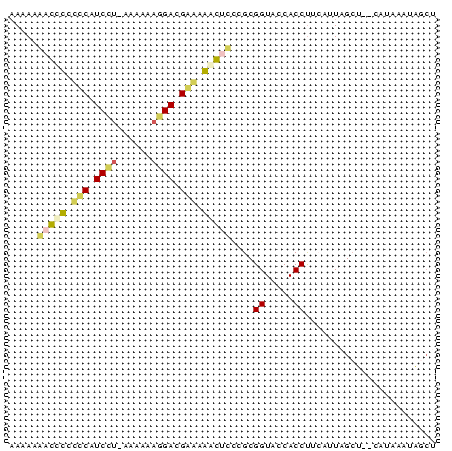
| Location | 3,133,268 – 3,133,343 |
|---|---|
| Length | 75 |
| Sequences | 6 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 68.13 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -25.33 |
| Energy contribution | -22.78 |
| Covariance contribution | -2.55 |
| Combinations/Pair | 1.74 |
| Mean z-score | -5.12 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 3133268 75 + 5227293 AGCUAUUCAUGAUAGCUAAUUAAGGUGGUACCGCGGGAGUCUCUCGUCCUUUUUUAAGGAUGAGGGGCUCUUUUU (((((((...))))))).......(((....)))(((((((((((((((((....)))))))))))))))))... ( -34.00) >Bcere.0 3157045 74 + 5224283 AGCUAUAAA-GAUAGCUAAUUAAGGUGGUACCGCGGGAGUCUCUCGUCCUUUUUUAAGGAUGAGGGACUCUUUUU ((((((...-.)))))).......(((....)))(((((((((((((((((....)))))))))))))))))... ( -34.50) >Bhalo.0 2322309 72 + 4202352 GGUUAUUUAUUG-ACC-AAUGAAGGUGGUACCGCGGG-CUCUCCCGUCCUUUUCUAAGGAUGGGGGAGCUUUUUU (((((.....))-)))-.......(((....)))(((-(((((((((((((....)))))))))))))))).... ( -35.20) >Bclau.0 3999426 72 - 4303871 GCUUUUUUGUGG-AGCGAAUGAAGGUGGCACCGCGGG-GUUAUCCGUCCUUUUUU-AGGAUGGGUAGCCCUUUUU ........((((-.((.(.......).)).))))(((-((((((((((((.....-))))))))))))))).... ( -32.30) >Blich.0 1756018 71 - 4222334 AGCAU-GUAUA--UGCUAAUGAAGGUGGUACCGCGAAACCUUUUCGUCCUUUGCG-AAGACGGAAAGGUUUUUUU (((((-....)--)))).......(((....)))((((((((((((((.......-..))))))))))))))... ( -23.20) >Bsubt.0 1742314 70 - 4214630 AGCACAAUUUG--UGCUAAUGAGGGUGGUACCGCG-AACCUUUUCGUCCUUUACG--UGAUGAAAAGGUUUUUUG (((((.....)--)))).......(((....)))(-(((((((((((((.....)--.))))))))))))).... ( -24.40) >consensus AGCUAUUUAUG__AGCUAAUGAAGGUGGUACCGCGGGACUCUCUCGUCCUUUUCU_AGGAUGAGGAGCUCUUUUU ........................(((....)))((((((((((((((((......))))))))))))))))... (-25.33 = -22.78 + -2.55)
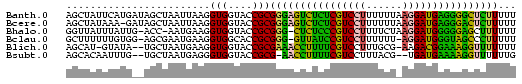
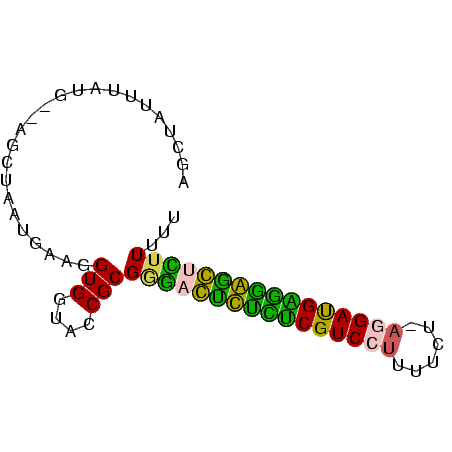
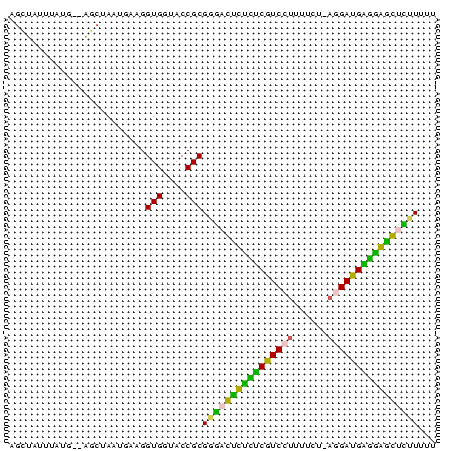
| Location | 3,133,268 – 3,133,343 |
|---|---|
| Length | 75 |
| Sequences | 6 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 68.13 |
| Mean single sequence MFE | -18.40 |
| Consensus MFE | -10.88 |
| Energy contribution | -10.05 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 3133268 75 + 5227293/0-75 AAAAAGAGCCCCUCAUCCUUAAAAAAGGACGAGAGACUCCCGCGGUACCACCUUAAUUAGCUAUCAUGAAUAGCU .....(((.(.(((.(((((....))))).))).).)))....((.....))......((((((.....)))))) ( -17.20) >Bcere.0 3157045 74 + 5224283/0-75 AAAAAGAGUCCCUCAUCCUUAAAAAAGGACGAGAGACUCCCGCGGUACCACCUUAAUUAGCUAUC-UUUAUAGCU .....(((((.(((.(((((....))))).))).)))))....((.....))......((((((.-...)))))) ( -22.00) >Bhalo.0 2322309 72 + 4202352/0-75 AAAAAAGCUCCCCCAUCCUUAGAAAAGGACGGGAGAG-CCCGCGGUACCACCUUCAUU-GGU-CAAUAAAUAACC ......((((.(((.(((((....))))).))).)))-).....(.((((.......)-)))-)........... ( -19.90) >Bclau.0 3999426 72 - 4303871/0-75 AAAAAGGGCUACCCAUCCU-AAAAAAGGACGGAUAAC-CCCGCGGUGCCACCUUCAUUCGCU-CCACAAAAAAGC .....(((.((.((.((((-.....)))).)).)).)-)).(.((.((...........)).-)).)........ ( -15.30) >Blich.0 1756018 71 - 4222334/0-75 AAAAAAACCUUUCCGUCUU-CGCAAAGGACGAAAAGGUUUCGCGGUACCACCUUCAUUAGCA--UAUAC-AUGCU ....((((((((.((((((-.....)))))).))))))))...((.....))......((((--(....-))))) ( -18.50) >Bsubt.0 1742314 70 - 4214630/0-75 CAAAAAACCUUUUCAUCA--CGUAAAGGACGAAAAGGUU-CGCGGUACCACCCUCAUUAGCA--CAAAUUGUGCU .....(((((((((.((.--(.....))).)))))))))-.(.((......)).)...((((--(.....))))) ( -17.50) >consensus AAAAAAACCCCCCCAUCCU_AAAAAAGGACGAAAAACUCCCGCGGUACCACCUUCAUUAGCU__CAUAAAUAGCU .....(((((.(((.((((......)))).))).)))))....((.....))....................... (-10.88 = -10.05 + -0.83)

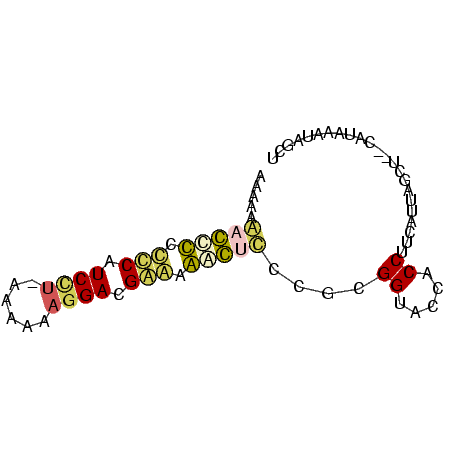

| Location | 3,133,268 – 3,133,343 |
|---|---|
| Length | 75 |
| Sequences | 6 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 68.13 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -25.33 |
| Energy contribution | -22.78 |
| Covariance contribution | -2.55 |
| Combinations/Pair | 1.74 |
| Mean z-score | -5.12 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 3133268 75 + 5227293/0-75 AGCUAUUCAUGAUAGCUAAUUAAGGUGGUACCGCGGGAGUCUCUCGUCCUUUUUUAAGGAUGAGGGGCUCUUUUU (((((((...))))))).......(((....)))(((((((((((((((((....)))))))))))))))))... ( -34.00) >Bcere.0 3157045 74 + 5224283/0-75 AGCUAUAAA-GAUAGCUAAUUAAGGUGGUACCGCGGGAGUCUCUCGUCCUUUUUUAAGGAUGAGGGACUCUUUUU ((((((...-.)))))).......(((....)))(((((((((((((((((....)))))))))))))))))... ( -34.50) >Bhalo.0 2322309 72 + 4202352/0-75 GGUUAUUUAUUG-ACC-AAUGAAGGUGGUACCGCGGG-CUCUCCCGUCCUUUUCUAAGGAUGGGGGAGCUUUUUU (((((.....))-)))-.......(((....)))(((-(((((((((((((....)))))))))))))))).... ( -35.20) >Bclau.0 3999426 72 - 4303871/0-75 GCUUUUUUGUGG-AGCGAAUGAAGGUGGCACCGCGGG-GUUAUCCGUCCUUUUUU-AGGAUGGGUAGCCCUUUUU ........((((-.((.(.......).)).))))(((-((((((((((((.....-))))))))))))))).... ( -32.30) >Blich.0 1756018 71 - 4222334/0-75 AGCAU-GUAUA--UGCUAAUGAAGGUGGUACCGCGAAACCUUUUCGUCCUUUGCG-AAGACGGAAAGGUUUUUUU (((((-....)--)))).......(((....)))((((((((((((((.......-..))))))))))))))... ( -23.20) >Bsubt.0 1742314 70 - 4214630/0-75 AGCACAAUUUG--UGCUAAUGAGGGUGGUACCGCG-AACCUUUUCGUCCUUUACG--UGAUGAAAAGGUUUUUUG (((((.....)--)))).......(((....)))(-(((((((((((((.....)--.))))))))))))).... ( -24.40) >consensus AGCUAUUUAUG__AGCUAAUGAAGGUGGUACCGCGGGACUCUCUCGUCCUUUUCU_AGGAUGAGGAGCUCUUUUU ........................(((....)))((((((((((((((((......))))))))))))))))... (-25.33 = -22.78 + -2.55)
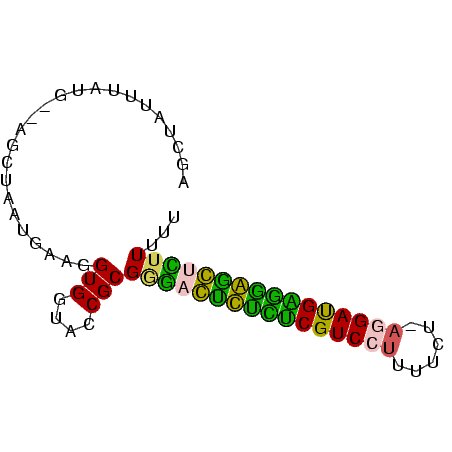
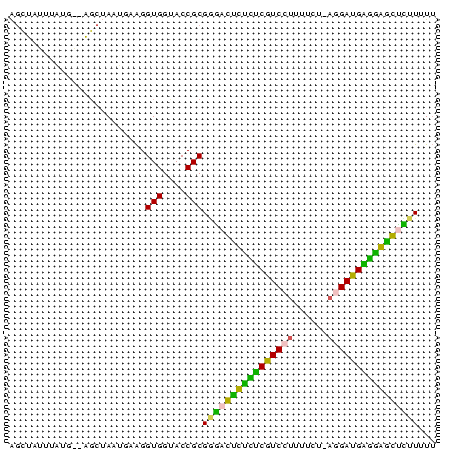
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:28:10 2006