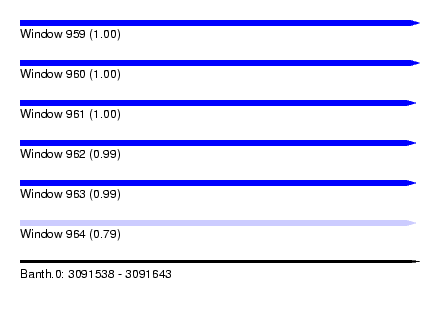
| Sequence ID | Banth.0 |
|---|---|
| Location | 3,091,538 – 3,091,643 |
| Length | 105 |
| Max. P | 0.999763 |

| Location | 3,091,538 – 3,091,643 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.45 |
| Mean single sequence MFE | -36.47 |
| Consensus MFE | -17.39 |
| Energy contribution | -17.45 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.34 |
| Mean z-score | -4.07 |
| Structure conservation index | 0.48 |
| SVM decision value | 3.65 |
| SVM RNA-class probability | 0.999489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 3091538 105 + 5227293/0-120 AAAAAACCUCUCCGAUAAGAAGAAGUUACGUAUAUAUCUUCGUCUUAUCUCCCAGAACAAAUUUGUUCUGCUGGAAUUAGCACCUGCAUC-GC--------------GGUUGCUGGGUUU ...(((((..((((((((((.((((.((......)).)))).)))))))...(((((((....)))))))..)))..(((((.((((...-))--------------)).)))))))))) ( -34.30) >Bcere.0 3108512 105 + 5224283/0-120 AAAAAACCUCUCCGAUAAGAAGAAGUUACGUAUAUAUCUUCGUCUUAUCUCCCAGAACAAAUAUGUUCUGCUGGAAUUAGCACCUGCAUC-GC--------------GGUUGCUGGGUUU ...(((((..((((((((((.((((.((......)).)))).)))))))...(((((((....)))))))..)))..(((((.((((...-))--------------)).)))))))))) ( -34.30) >Bhalo.0 2853388 114 - 4202352/0-120 AAAAAACCUUUCCGAUAAGGAAAGGUU-----UUCGCCUUCG-CUUAUCUUCCAGAAUGAAAUCAUUCUGCUGAAAUUAGCACCUUCGUUUGCUUGGCUAACAAACGGGUUGCCGGGCAU ..(((((((((((.....)))))))))-----)).......(-((..((...(((((((....)))))))..))....))).........((((((((.(((......))))))))))). ( -40.00) >Bclau.0 3352324 104 - 4303871/0-120 AAAGAAGCU-----GCGAAAAUCCGUU---UCCGUUUUUCCGGCUUAUCUUUCAGAAUGAUCCCAUUCUGCAGGAAUUGGCACCUUCCCC-GU-------AACGGGAGGUUGCUGGACGU ....(((((-----(.(((((..((..---..)).))))))))))).((((.(((((((....))))))).)))).(..(((.((((((.-..-------...)))))).)))..).... ( -37.30) >consensus AAAAAACCUCUCCGAUAAGAAGAAGUU___UAUAUAUCUUCGUCUUAUCUCCCAGAACAAAUUCAUUCUGCUGGAAUUAGCACCUGCAUC_GC______________GGUUGCUGGGCUU .............(((((((.((((............)))).)))))))((((((((((....)))))))..))).((((((((.......................)).)))))).... (-17.39 = -17.45 + 0.06)

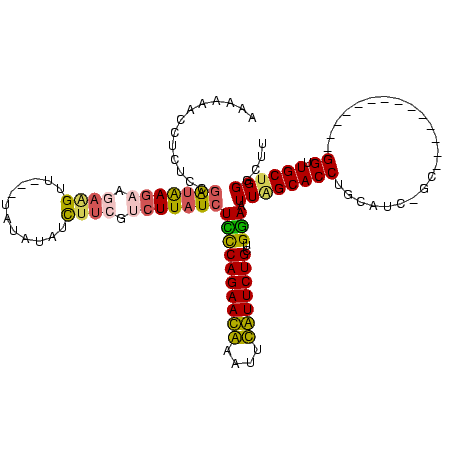
| Location | 3,091,538 – 3,091,643 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.45 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -16.38 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.45 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 3091538 105 + 5227293/0-120 AAACCCAGCAACC--------------GC-GAUGCAGGUGCUAAUUCCAGCAGAACAAAUUUGUUCUGGGAGAUAAGACGAAGAUAUAUACGUAACUUCUUCUUAUCGGAGAGGUUUUUU (((((.((((.((--------------..-......))))))..((((..(((((((....)))))))...(((((((.((((.(((....))).)))).))))))))))).)))))... ( -34.70) >Bcere.0 3108512 105 + 5224283/0-120 AAACCCAGCAACC--------------GC-GAUGCAGGUGCUAAUUCCAGCAGAACAUAUUUGUUCUGGGAGAUAAGACGAAGAUAUAUACGUAACUUCUUCUUAUCGGAGAGGUUUUUU (((((.((((.((--------------..-......))))))..((((..(((((((....)))))))...(((((((.((((.(((....))).)))).))))))))))).)))))... ( -35.50) >Bhalo.0 2853388 114 - 4202352/0-120 AUGCCCGGCAACCCGUUUGUUAGCCAAGCAAACGAAGGUGCUAAUUUCAGCAGAAUGAUUUCAUUCUGGAAGAUAAG-CGAAGGCGAA-----AACCUUUCCUUAUCGGAAAGGUUUUUU ..(((.....(((((((((((.....))))))))..)))(((.((((...(((((((....)))))))..)))).))-)...)))(((-----(((((((((.....)))))))))))). ( -42.00) >Bclau.0 3352324 104 - 4303871/0-120 ACGUCCAGCAACCUCCCGUU-------AC-GGGGAAGGUGCCAAUUCCUGCAGAAUGGGAUCAUUCUGAAAGAUAAGCCGGAAAAACGGA---AACGGAUUUUCGC-----AGCUUCUUU .......(((.((((((...-------..-.))).)))))).........(((((((....)))))))(((((..(((((((((..((..---..))..)))))).-----.)))))))) ( -33.50) >consensus AAACCCAGCAACC______________GC_GAUGAAGGUGCUAAUUCCAGCAGAACAAAUUCAUUCUGGAAGAUAAGACGAAGAUAUAUA___AACUUCUUCUUAUCGGAGAGGUUUUUU ......((((.((.......................))))))..((((..(((((((....))))))))))).....................(((((((((.....))))))))).... (-16.38 = -17.00 + 0.62)

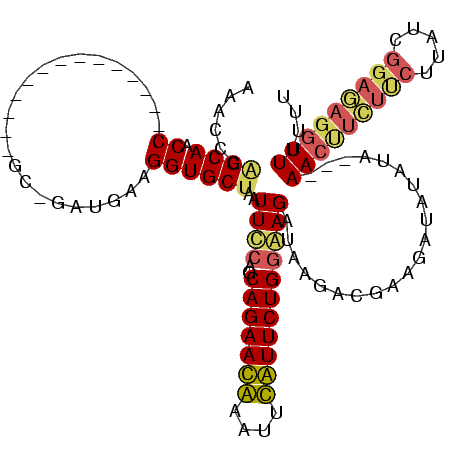
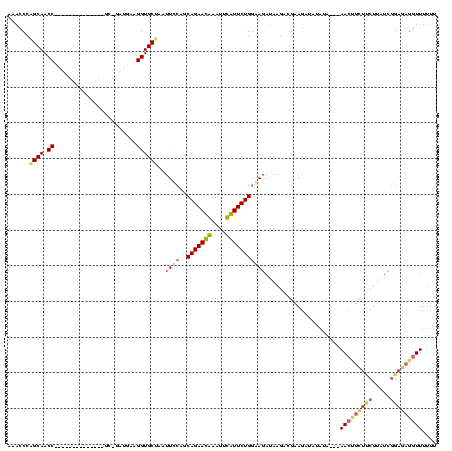
| Location | 3,091,538 – 3,091,642 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.57 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -25.38 |
| Energy contribution | -23.00 |
| Covariance contribution | -2.38 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.74 |
| SVM decision value | 4.03 |
| SVM RNA-class probability | 0.999763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 3091538 104 + 5227293/40-160 CGUCUUAUCUCCCAGAACAAAUUUGUUCUGCUGGAAUUAGCACCUGCAUC-GC--------------GGUUGCUGGGUUUCAUCGGGCC-AGUCCCUCCACCUCUCUGGAUAAGAAAAUA ..((((((((..(((((((....))))))).((((.((((((.((((...-))--------------)).))))))........(((..-...))))))).......))))))))..... ( -31.70) >Bcere.0 3108512 104 + 5224283/40-160 CGUCUUAUCUCCCAGAACAAAUAUGUUCUGCUGGAAUUAGCACCUGCAUC-GC--------------GGUUGCUGGGUUUCAUCGGGCC-AGUCCCUCCACCUCUCUGGAUAAGAAAAUA ..((((((((..(((((((....))))))).((((.((((((.((((...-))--------------)).))))))........(((..-...))))))).......))))))))..... ( -31.70) >Bhalo.0 2853388 116 - 4202352/40-160 CG-CUUAUCUUCCAGAAUGAAAUCAUUCUGCUGAAAUUAGCACCUUCGUUUGCUUGGCUAACAAACGGGUUGCCGGGCAUCAUCGGGUC-AGUCCCUCUGCCUCUCUCGAUAAGUC--UU .(-((((((((((((((((....)))))))..)))....(((........((((((((.(((......))))))))))).....(((..-...)))..))).......))))))).--.. ( -35.10) >Bclau.0 3352324 112 - 4303871/40-160 CGGCUUAUCUUUCAGAAUGAUCCCAUUCUGCAGGAAUUGGCACCUUCCCC-GU-------AACGGGAGGUUGCUGGACGUCAUUGGGCCUAAUCCCUCAGUCGCUCUUGAUAAGUCCUUU .((((((((...(((((((....))))))).((((.(..(((.((((((.-..-------...)))))).)))..)..((.((((((........)))))).)))))))))))))).... ( -38.80) >consensus CGUCUUAUCUCCCAGAACAAAUUCAUUCUGCUGGAAUUAGCACCUGCAUC_GC______________GGUUGCUGGGCUUCAUCGGGCC_AGUCCCUCCACCUCUCUGGAUAAGAAAAUA ..(((((((((((((((((....)))))))..)))..(((((((.......................)).)))))(((......(((......)))...)))......)))))))..... (-25.38 = -23.00 + -2.38)

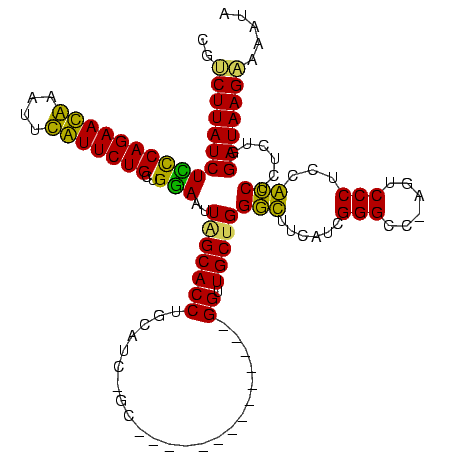
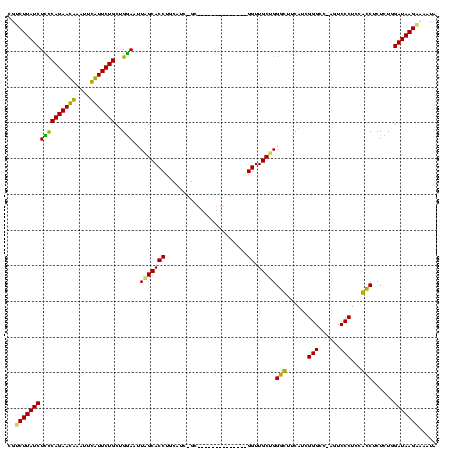
| Location | 3,091,538 – 3,091,642 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.57 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -24.08 |
| Energy contribution | -22.32 |
| Covariance contribution | -1.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 3091538 104 + 5227293/40-160 UAUUUUCUUAUCCAGAGAGGUGGAGGGACU-GGCCCGAUGAAACCCAGCAACC--------------GC-GAUGCAGGUGCUAAUUCCAGCAGAACAAAUUUGUUCUGGGAGAUAAGACG .....((((((((.......(((((((...-..)))..........((((.((--------------..-......))))))...)))).(((((((....))))))).).))))))).. ( -29.80) >Bcere.0 3108512 104 + 5224283/40-160 UAUUUUCUUAUCCAGAGAGGUGGAGGGACU-GGCCCGAUGAAACCCAGCAACC--------------GC-GAUGCAGGUGCUAAUUCCAGCAGAACAUAUUUGUUCUGGGAGAUAAGACG .....((((((((.......(((((((...-..)))..........((((.((--------------..-......))))))...)))).(((((((....))))))).).))))))).. ( -30.60) >Bhalo.0 2853388 116 - 4202352/40-160 AA--GACUUAUCGAGAGAGGCAGAGGGACU-GACCCGAUGAUGCCCGGCAACCCGUUUGUUAGCCAAGCAAACGAAGGUGCUAAUUUCAGCAGAAUGAUUUCAUUCUGGAAGAUAAG-CG ..--(.(((((((((((.((((..(((...-..))).....)))))((((.((((((((((.....))))))))..))))))..))))..(((((((....)))))))...))))))-). ( -41.20) >Bclau.0 3352324 112 - 4303871/40-160 AAAGGACUUAUCAAGAGCGACUGAGGGAUUAGGCCCAAUGACGUCCAGCAACCUCCCGUU-------AC-GGGGAAGGUGCCAAUUCCUGCAGAAUGGGAUCAUUCUGAAAGAUAAGCCG ...((.((((((...........(((((((.(((((..((.(.....)))...((((...-------..-.)))).)).)))))))))).(((((((....)))))))...)))))))). ( -37.90) >consensus AAUUGACUUAUCCAGAGAGGCGGAGGGACU_GGCCCGAUGAAACCCAGCAACC______________GC_GAUGAAGGUGCUAAUUCCAGCAGAACAAAUUCAUUCUGGAAGAUAAGACG ......((((((....(.(((...(((......)))......))))((((.((.......................))))))........(((((((....)))))))...))))))... (-24.08 = -22.32 + -1.75)

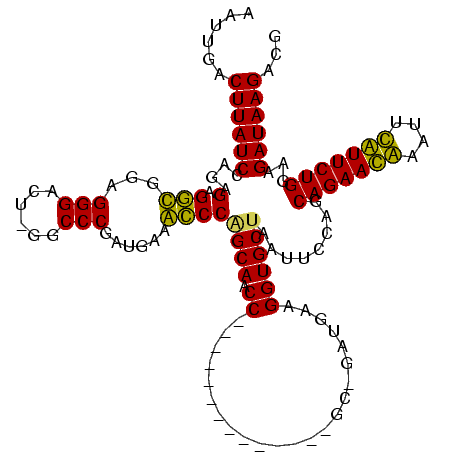
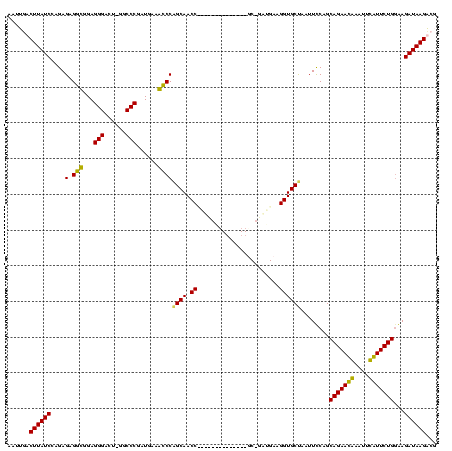
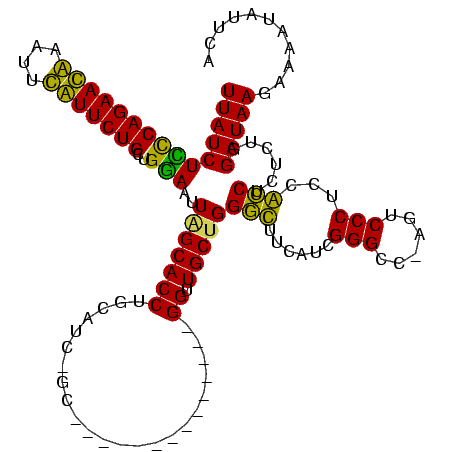
| Location | 3,091,538 – 3,091,642 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.72 |
| Mean single sequence MFE | -30.53 |
| Consensus MFE | -21.62 |
| Energy contribution | -19.25 |
| Covariance contribution | -2.38 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 3091538 104 + 5227293/44-164 UUAUCUCCCAGAACAAAUUUGUUCUGCUGGAAUUAGCACCUGCAUC-GC--------------GGUUGCUGGGUUUCAUCGGGCC-AGUCCCUCCACCUCUCUGGAUAAGAAAAUAUUCA ...(((..(((((((....)))))))..))).((((((.((((...-))--------------)).)))))).((((...(((..-...)))((((......))))...))))....... ( -27.60) >Bcere.0 3108512 104 + 5224283/44-164 UUAUCUCCCAGAACAAAUAUGUUCUGCUGGAAUUAGCACCUGCAUC-GC--------------GGUUGCUGGGUUUCAUCGGGCC-AGUCCCUCCACCUCUCUGGAUAAGAAAAUAUUCA ...(((..(((((((....)))))))..))).((((((.((((...-))--------------)).)))))).((((...(((..-...)))((((......))))...))))....... ( -27.60) >Bhalo.0 2853388 117 - 4202352/44-164 UUAUCUUCCAGAAUGAAAUCAUUCUGCUGAAAUUAGCACCUUCGUUUGCUUGGCUAACAAACGGGUUGCCGGGCAUCAUCGGGUC-AGUCCCUCUGCCUCUCUCGAUAAGUC--UUUUUA (((((((((((((((....)))))))..)))....(((........((((((((.(((......))))))))))).....(((..-...)))..))).......)))))...--...... ( -30.90) >Bclau.0 3352324 112 - 4303871/44-164 UUAUCUUUCAGAAUGAUCCCAUUCUGCAGGAAUUGGCACCUUCCCC-GU-------AACGGGAGGUUGCUGGACGUCAUUGGGCCUAAUCCCUCAGUCGCUCUUGAUAAGUCCUUUUUUA ...((((.(((((((....))))))).))))...((((.((((((.-..-------...)))))).))))((((((((..((((..............)))).))))..))))....... ( -36.04) >consensus UUAUCUCCCAGAACAAAUUCAUUCUGCUGGAAUUAGCACCUGCAUC_GC______________GGUUGCUGGGCUUCAUCGGGCC_AGUCCCUCCACCUCUCUGGAUAAGAAAAUAUUCA (((((((((((((((....)))))))..)))..(((((((.......................)).)))))(((......(((......)))...)))......)))))........... (-21.62 = -19.25 + -2.38)

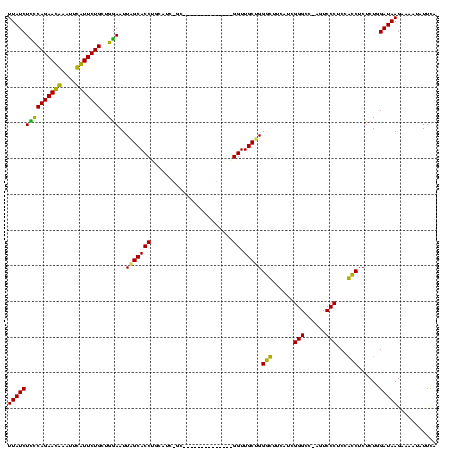
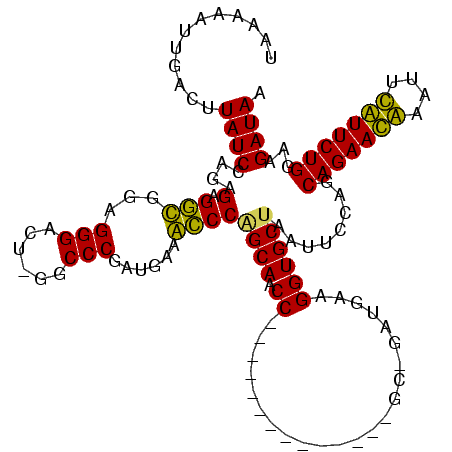
| Location | 3,091,538 – 3,091,642 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.72 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -20.62 |
| Energy contribution | -18.88 |
| Covariance contribution | -1.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 3091538 104 + 5227293/44-164 UGAAUAUUUUCUUAUCCAGAGAGGUGGAGGGACU-GGCCCGAUGAAACCCAGCAACC--------------GC-GAUGCAGGUGCUAAUUCCAGCAGAACAAAUUUGUUCUGGGAGAUAA ....(((((((...((((......))))((((.(-(((((..((.....))(((...--------------..-..))).)).)))).))))..(((((((....)))))))))))))). ( -28.40) >Bcere.0 3108512 104 + 5224283/44-164 UGAAUAUUUUCUUAUCCAGAGAGGUGGAGGGACU-GGCCCGAUGAAACCCAGCAACC--------------GC-GAUGCAGGUGCUAAUUCCAGCAGAACAUAUUUGUUCUGGGAGAUAA ....(((((((...((((......))))((((.(-(((((..((.....))(((...--------------..-..))).)).)))).))))..(((((((....)))))))))))))). ( -29.20) >Bhalo.0 2853388 117 - 4202352/44-164 UAAAAA--GACUUAUCGAGAGAGGCAGAGGGACU-GACCCGAUGAUGCCCGGCAACCCGUUUGUUAGCCAAGCAAACGAAGGUGCUAAUUUCAGCAGAAUGAUUUCAUUCUGGAAGAUAA ......--....(((((((((.((((..(((...-..))).....)))))((((.((((((((((.....))))))))..))))))..))))..(((((((....)))))))...)))). ( -37.20) >Bclau.0 3352324 112 - 4303871/44-164 UAAAAAAGGACUUAUCAAGAGCGACUGAGGGAUUAGGCCCAAUGACGUCCAGCAACCUCCCGUU-------AC-GGGGAAGGUGCCAAUUCCUGCAGAAUGGGAUCAUUCUGAAAGAUAA .......((((...(((........)))(((......)))......)))).(((.((((((...-------..-.))).))))))....((...(((((((....)))))))...))... ( -30.90) >consensus UAAAAAUUGACUUAUCCAGAGAGGCGGAGGGACU_GGCCCGAUGAAACCCAGCAACC______________GC_GAUGAAGGUGCUAAUUCCAGCAGAACAAAUUCAUUCUGGAAGAUAA ............((((....(.(((...(((......)))......))))((((.((.......................))))))........(((((((....)))))))...)))). (-20.62 = -18.88 + -1.75)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:28:08 2006