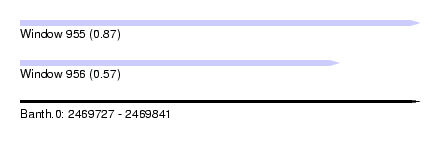
| Sequence ID | Banth.0 |
|---|---|
| Location | 2,469,727 – 2,469,841 |
| Length | 114 |
| Max. P | 0.873886 |

| Location | 2,469,727 – 2,469,841 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.27 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -22.98 |
| Energy contribution | -19.44 |
| Covariance contribution | -3.54 |
| Combinations/Pair | 1.41 |
| Mean z-score | -0.53 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
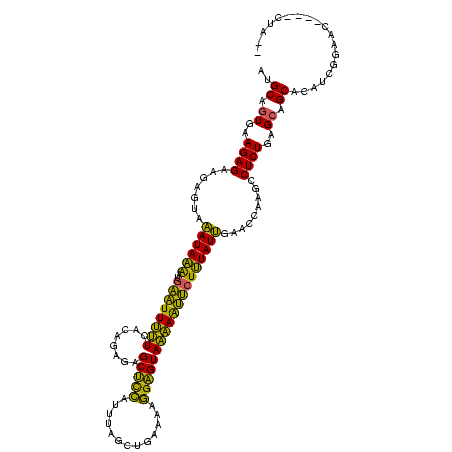
>Banth.0 2469727 114 + 5227293/0-120 AUGCAGUGAAGAGAAGAGUAAAUAAGAUGAAUUUUUCACAGAGAGCUCCAUUUAGCUGAAAAGGAGUAAAAAUUCUUUAUUGAACCAAGCCUCUGAGCAGCGCAUCGGAAU----CUA-- ...((((((((((((((((...........))))))).......(((((.(((.....))).)))))......))))))))).....((..(((((((...)).)))))..----)).-- ( -23.60) >Bcere.0 2551778 114 + 5224283/0-120 AUGCAGUGAAGAGAAGAGUAAAUAAAAUGAAUUUUUCACAGAGAGCUCCAUUUAGCUGAAAAGGAGUAAAAAUUCUUUAUUGAACCAAGCCUCUGAGCAGCACAUCGGAAC----CUA-- ...((((((((((((((((...........))))))).......(((((.(((.....))).)))))......))))))))).....((..(((((........)))))..----)).-- ( -23.20) >Bhalo.0 3321532 120 - 4202352/0-120 ACGCGAUGAAGAGAAGAGUACAUACAUCCCUUUCCUCACAGAAAGCCUGUAGUUGCUGAGAACGGGUAGGAGAGCUGUAUGGAAGCAAGACUCUGAGCGGCACAUCGGAACAACCCUACG ...(((((..(...(((((.((((((...(((((((........((((((...........))))))))))))).))))))........)))))...)....)))))............. ( -28.90) >consensus AUGCAGUGAAGAGAAGAGUAAAUAAAAUGAAUUUUUCACAGAGAGCUCCAUUUAGCUGAAAAGGAGUAAAAAUUCUUUAUUGAACCAAGCCUCUGAGCAGCACAUCGGAAC____CUA__ ..((.((..((((.......((((((..((((((((........(((((.............))))))))))))))))))).........))))..)).))................... (-22.98 = -19.44 + -3.54)


| Location | 2,469,727 – 2,469,818 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.28 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -15.34 |
| Energy contribution | -14.68 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
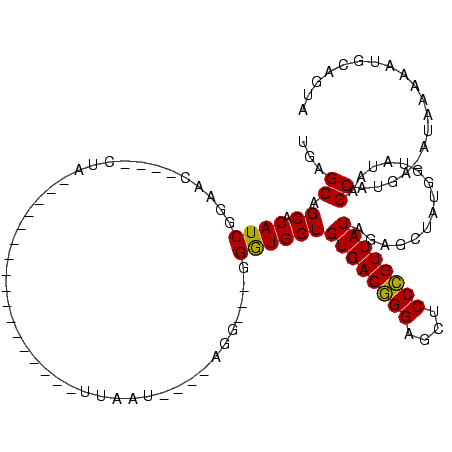
>Banth.0 2469727 91 + 5227293/93-213 UGAGCAGCGCAUCGGAAU----CUA-----------------UUAAU----AGA---GGGUGGUGUGACGGGAGCUCCCGUUAUAGAGCUAUGGUAUAAGCAUGA-AUGAAAAUGCAGUA ..(((..(((((((...(----(((-----------------....)----)))---...)))))))(((((....)))))......))).........((((..-......)))).... ( -25.20) >Bcere.0 2551778 91 + 5224283/93-213 UGAGCAGCACAUCGGAAC----CUA-----------------UUAUU----AGG---GGGUGGUGUGACGGGAGCUCCCGUUAUAGAGCUAUGGUAUAAGCAUGA-AUUAGAAUGCAGUA ..(((..(((((((...(----(((-----------------....)----)))---...)))))))(((((....)))))......))).........((((..-......)))).... ( -28.00) >Bhalo.0 3321532 119 - 4202352/93-213 UGAGCGGCACAUCGGAACAACCCUACGCCGGCCGGCAAGCAUUUAAUCCAAAGGUUGGGAUGGUGUGACAGGAGCUCCUGUUAUUGAGCAAUG-UAUAGGCUCACUAUACACAUGAGGUU ...((..(((((((...(((((....(((....))).....(((.....))))))))...)))))))(((((....)))))......))..((-(((((.....)))))))......... ( -34.60) >consensus UGAGCAGCACAUCGGAAC____CUA_________________UUAAU____AGG___GGGUGGUGUGACGGGAGCUCCCGUUAUAGAGCUAUGGUAUAAGCAUGA_AUAAAAAUGCAGUA ...((.((.((((.............................................))))))((((((((....))))))))...............))................... (-15.34 = -14.68 + -0.66)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:28:02 2006