
| Sequence ID | Banth.0 |
|---|---|
| Location | 33,995 – 34,115 |
| Length | 120 |
| Max. P | 0.983138 |

| Location | 33,995 – 34,115 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.72 |
| Mean single sequence MFE | -46.04 |
| Consensus MFE | -45.31 |
| Energy contribution | -41.73 |
| Covariance contribution | -3.58 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
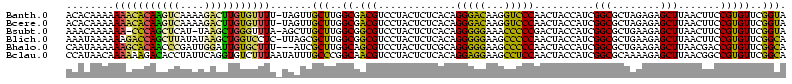
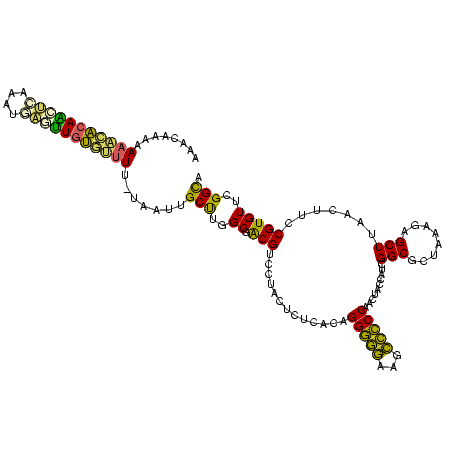
>Banth.0 33995 120 + 5227293/0-120 GUUUGGUGAUGAUGGCAGAGAGGUCACACCCGUUCCCAUACCGAACACGGAAGUUAAGCUCUCUAGCGCCGAUGGUAGUUGGGACCUUGUCCCUGUGAGAGUAGGACGUCGCCAAGCAAC (((((((((((((((..(((.((......)).))))))).((....((....))...((((((.((((((...))).)))(((((...)))))...)))))).)).)))))))))))... ( -42.70) >Bcere.0 33975 120 + 5224283/0-120 GUUUGGUGAUGAUGGCAGAGAGGUCACACCCGUUCCCAUACCGAACACGGAAGUUAAGCUCUCUAGCGCCGAUGGUAGUUGGGACCUUGUCCCUGUGAGAGUAGGACGUCGCCAAGCAAC (((((((((((((((..(((.((......)).))))))).((....((....))...((((((.((((((...))).)))(((((...)))))...)))))).)).)))))))))))... ( -42.70) >Bsubt.0 35233 120 + 4214630/0-120 GUUUGGUGGCGAUAGCGAAGAGGUCACACCCGUUCCCAUACCGAACACGGAAGUUAAGCUCUUCAGCGCCGAUGGUAGUCGGGGGUUUCCCCCUGUGAGAGUAGGACGCCGCCAAGCAAG (((((((((((...((((((((.........((((.......))))((....))....)))))).)).(((((....)))(((((...)))))..........)).)))))))))))... ( -47.90) >Blich.0 39551 120 + 4222334/0-120 GUUUGGUGGCGAUAGCGAAGAGGUCACACCCGUUCCCAUGCCGAACACGGAAGUUAAGCUCUUCAGCGCCGAUGGUAGUUGGGGGCUUCCCCCUGUGAGAGUAGGACGCCGCCAAGCGCU (((((((((((...((((((((.........((((.......))))((....))....)))))).)).((..........(((((...)))))..........)).)))))))))))... ( -46.65) >Bhalo.0 28399 120 + 4202352/0-120 GUUUGGUGGCGAUAGCGAAGAGGCCACACCCGUUCCCAUGCCGAACACGGUCGUUAAGCUCUUCAGCGCCGAUGGUAGUUGGGGGCUUCCCCCUGCGAGAGUAGGACGCUGCCAAGCGAU ((((((..(((........((((((...(((((((.......))))((.((((....(((....)))..)))).))....)))))))))..(((((....))))).)))..))))))... ( -51.00) >Bclau.0 16731 120 + 4303871/0-120 GUCCGGUGACAAUGGCAAAGAGGUCACACCCGUUCCCAUGCCGAACACGGCCGUUAAGCUCUUUUGCGCCGAUGGUAGUUGGAGGCUUCCUCCUGUGAGAGUAGGACGUUGCCGGGCAAA ((((((..((.((((....(.((.....)))....))))((((....)))).(((..((((((..(((((...)))....(((((...))))).))))))))..)))))..))))))... ( -45.30) >consensus GUUUGGUGACGAUAGCAAAGAGGUCACACCCGUUCCCAUACCGAACACGGAAGUUAAGCUCUUCAGCGCCGAUGGUAGUUGGGGGCUUCCCCCUGUGAGAGUAGGACGCCGCCAAGCAAC (((((((((((...((((((((.........((((.......))))((....))....)))))).)).((.((....(..(((((...)))))..)....)).)).)))))))))))... (-45.31 = -41.73 + -3.58)

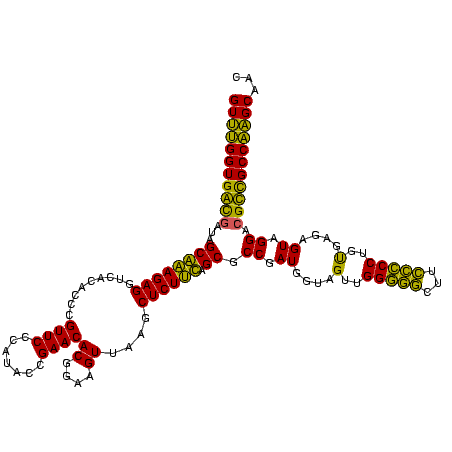
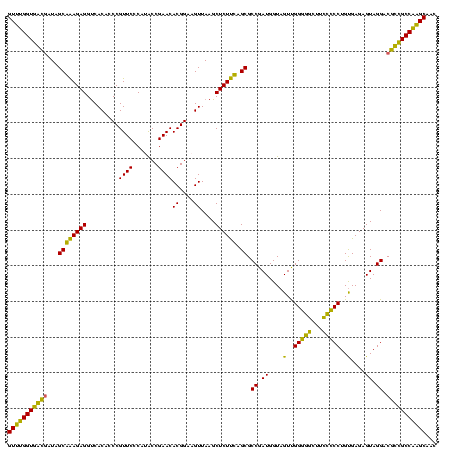
| Location | 33,995 – 34,114 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.82 |
| Mean single sequence MFE | -44.10 |
| Consensus MFE | -38.33 |
| Energy contribution | -34.45 |
| Covariance contribution | -3.88 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 33995 119 + 5227293/38-158 UACCGAACACGGAAGUUAAGCUCUCUAGCGCCGAUGGUAGUUGGGACCUUGUCCCUGUGAGAGUAGGACGUCGCCAAGCAACUA-AAAACACAAGUCUUUUGACUUGUGUUUUUUUGUGU ......((((((..(((..((((((.((((((...))).)))(((((...)))))...))))))..)))....))........(-((((((((((((....)))))))))))))..)))) ( -41.70) >Bcere.0 33975 119 + 5224283/38-158 UACCGAACACGGAAGUUAAGCUCUCUAGCGCCGAUGGUAGUUGGGACCUUGUCCCUGUGAGAGUAGGACGUCGCCAAGCAACUA-AAAACACAAGUCUUUUGACUUGUGUUUUUUUGUGU ......((((((..(((..((((((.((((((...))).)))(((((...)))))...))))))..)))....))........(-((((((((((((....)))))))))))))..)))) ( -41.70) >Bsubt.0 35233 117 + 4214630/38-158 UACCGAACACGGAAGUUAAGCUCUUCAGCGCCGAUGGUAGUCGGGGGUUUCCCCCUGUGAGAGUAGGACGCCGCCAAGCAAGCU-UAAACCCAGCUUA-AUGAGCUGGG-UUUUUUGUUU .........(((..(((..(((((((((..(((((....)))))(((....)))))).))))))..))).)))..(((((((..-.(((((((((((.-..))))))))-)))))))))) ( -42.70) >Blich.0 39551 119 + 4222334/38-158 UGCCGAACACGGAAGUUAAGCUCUUCAGCGCCGAUGGUAGUUGGGGGCUUCCCCCUGUGAGAGUAGGACGCCGCCAAGCGCUAA-GAGGACCAGCUUAUAUAAGCUGGUCUUUUUUAUUU ..(((....)))........(((((.(((((.........((((.(((.(((..((.....))..))).))).)))))))))))-)))(((((((((....))))))))).......... ( -46.10) >Bhalo.0 28399 117 + 4202352/38-158 UGCCGAACACGGUCGUUAAGCUCUUCAGCGCCGAUGGUAGUUGGGGGCUUCCCCCUGCGAGAGUAGGACGCUGCCAAGCGAU---AAAGCACAAUCCAAUCGGGUUGUGCUUUUUUAUUG .((((....)))).(..((((.((((((((((...))).)))))))))))..)(((((....))))).((((....))))..---((((((((((((....))))))))))))....... ( -47.60) >Bclau.0 16731 120 + 4303871/38-158 UGCCGAACACGGCCGUUAAGCUCUUUUGCGCCGAUGGUAGUUGGAGGCUUCCUCCUGUGAGAGUAGGACGUUGCCGGGCAAAUAUUAAAGACACCUGAAUAGGUGUCUUUUUUGUUAUGG ((((.....((((((((..((((((..(((((...)))....(((((...))))).))))))))..))))..))))))))((((..((((((((((....))))))))))..)))).... ( -44.80) >consensus UACCGAACACGGAAGUUAAGCUCUUCAGCGCCGAUGGUAGUUGGGGGCUUCCCCCUGUGAGAGUAGGACGCCGCCAAGCAACUA_AAAACACAACUCAUUUGAGUUGUGUUUUUUUAUGU ..(((....)))..(((..((((((..(((((...)))....(((((...))))).))))))))..)))((......))......((((((((((((....))))))))))))....... (-38.33 = -34.45 + -3.88)

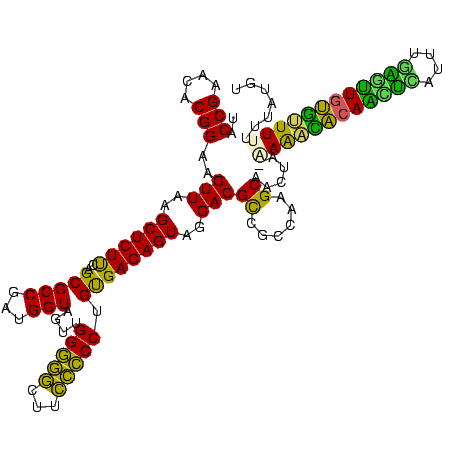
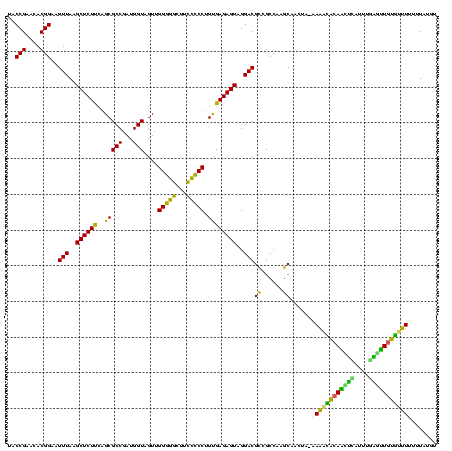
| Location | 33,995 – 34,114 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.82 |
| Mean single sequence MFE | -40.65 |
| Consensus MFE | -30.62 |
| Energy contribution | -28.10 |
| Covariance contribution | -2.52 |
| Combinations/Pair | 1.52 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 33995 119 + 5227293/38-158 ACACAAAAAAACACAAGUCAAAAGACUUGUGUUUU-UAGUUGCUUGGCGACGUCCUACUCUCACAGGGACAAGGUCCCAACUACCAUCGGCGCUAGAGAGCUUAACUUCCGUGUUCGGUA ((((..(((((((((((((....))))))))))))-).(((((...)))))......(((((...(((((...))))).....((...)).....)))))..........))))...... ( -36.90) >Bcere.0 33975 119 + 5224283/38-158 ACACAAAAAAACACAAGUCAAAAGACUUGUGUUUU-UAGUUGCUUGGCGACGUCCUACUCUCACAGGGACAAGGUCCCAACUACCAUCGGCGCUAGAGAGCUUAACUUCCGUGUUCGGUA ((((..(((((((((((((....))))))))))))-).(((((...)))))......(((((...(((((...))))).....((...)).....)))))..........))))...... ( -36.90) >Bsubt.0 35233 117 + 4214630/38-158 AAACAAAAAA-CCCAGCUCAU-UAAGCUGGGUUUA-AGCUUGCUUGGCGGCGUCCUACUCUCACAGGGGGAAACCCCCGACUACCAUCGGCGCUGAAGAGCUUAACUUCCGUGUUCGGUA .......(((-(((((((...-..)))))))))).-.....((((..(((((((...........((((....))))...........)))))))..)))).......(((....))).. ( -45.65) >Blich.0 39551 119 + 4222334/38-158 AAAUAAAAAAGACCAGCUUAUAUAAGCUGGUCCUC-UUAGCGCUUGGCGGCGUCCUACUCUCACAGGGGGAAGCCCCCAACUACCAUCGGCGCUGAAGAGCUUAACUUCCGUGUUCGGCA ..........(((((((((....))))))))).((-(((((((((((.(((.(((..((.....))..))).))).))).........))))))).)))(((.(((......))).))). ( -45.20) >Bhalo.0 28399 117 + 4202352/38-158 CAAUAAAAAAGCACAACCCGAUUGGAUUGUGCUUU---AUCGCUUGGCAGCGUCCUACUCUCGCAGGGGGAAGCCCCCAACUACCAUCGGCGCUGAAGAGCUUAACGACCGUGUUCGGCA .......(((((((((.((....)).)))))))))---...((((..(((((((...........((((....))))...........)))))))..)))).....(.(((....)))). ( -42.45) >Bclau.0 16731 120 + 4303871/38-158 CCAUAACAAAAAAGACACCUAUUCAGGUGUCUUUAAUAUUUGCCCGGCAACGUCCUACUCUCACAGGAGGAAGCCUCCAACUACCAUCGGCGCAAAAGAGCUUAACGGCCGUGUUCGGCA ..........((((((((((....))))))))))....((((((((....)..............(((((...)))))..........)).)))))...........((((....)))). ( -36.80) >consensus AAACAAAAAAACACAACUCAAAUGAGUUGUGUUUU_UAAUUGCUUGGCGACGUCCUACUCUCACAGGGGGAAGCCCCCAACUACCAUCGGCGCUAAAGAGCUUAACUUCCGUGUUCGGCA ........(((((((((((....))))))))))).......(((..((.(((.............(((((...)))))..........(((........))).......)))))..))). (-30.62 = -28.10 + -2.52)
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:28:00 2006