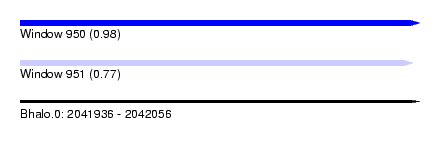
| Sequence ID | Bhalo.0 |
|---|---|
| Location | 2,041,936 – 2,042,056 |
| Length | 120 |
| Max. P | 0.983143 |

| Location | 2,041,936 – 2,042,056 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.78 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -34.29 |
| Energy contribution | -32.18 |
| Covariance contribution | -2.10 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
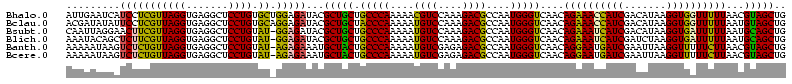
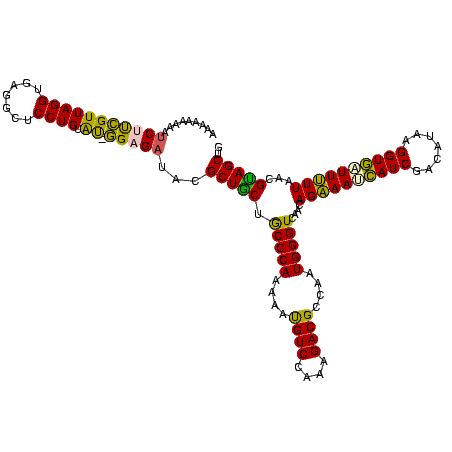
>Bhalo.0 2041936 120 + 4202352/0-120 AUUGAAUCAUCCUCGUUAGGUGAGGCUCCUGUGCUGGAGAUACGCUGCUGCCCAAAAACGUCCAAAGACGCCAAUGGGUCAACAGAAACCAUCGACAUAAGGUGGUUUUUAACGUAGCUG ..........(((((.....)))))((((......))))....(((((.(((((....((((....))))....)))))....((((((((((.......))))))))))...))))).. ( -39.00) >Bclau.0 3492933 120 - 4303871/0-120 ACGAUAUAUUCCUCGUUAGGUGAGGCUCCUGUGCAGGAGAUACGCUGCUACCCAAAAAUGUCCAAAGACGCCAAUGGGUCAACAGAAACCAUCGACAUAAGGUGGUUUUUAAUGUAGCUG ..........(((((.....)))))(((((....)))))....(((((.(((((....((((....))))....)))))....((((((((((.......))))))))))...))))).. ( -37.80) >Bsubt.0 1394917 119 + 4214630/0-120 CAAUUAGGAACUUCGUUAGGUGAGGCUCCUGUAU-GGAGAUACGCUGCUGCCCAAAAAUGUCCAAAGACGCCAAUGGGUCAACAGAAAUCAUCGACAUAAGGUGAUUUUUAAUGCAGCUG .....((((.(((((.....))))).))))((((-....))))(((((.(((((....((((....))))....)))))....((((((((((.......))))))))))...))))).. ( -36.60) >Blich.0 1419296 119 + 4222334/0-120 AAAUACAGCUCUUCGUUAGGUGAGGCUCCUGUAU-GGAGAUACGCUGCUGCCCAAAAAUGUCCAAAGACGCCAAUGGGUCAACAGAAAUCAUCGAUCUAAGGUGAUUUUUAAUGCAGCUG .........(((((((((((.......)))).))-)))))...(((((.(((((....((((....))))....)))))....((((((((((.......))))))))))...))))).. ( -36.60) >Banth.0 1510394 119 + 5227293/0-120 AAAAAUAAGUCUCUGUUAGGUGAGGCUCCUGUAU-AGAGAAAUGCUACUGCCCAAAAAUGUCGAGAGACGCCAAUGGGUCAACAGGAAUGAUCGAAUUAAGGUUUUUCUUAACGUAGCUG .........(((((((((((.......)))).))-)))))...(((((.(((((....((((....))))....)))))....(((((.((((.......)))).)))))...))))).. ( -35.70) >Bcere.0 1660585 119 + 5224283/0-120 AAAAAUAAGUCUCUGUUAGGUGAGGCUCCUGUAU-AGAGAAAUGCUACUGCCCAAAAAUGUCGAGAGACGCCAAUGGGUCAACAGGAAUGAUCGAAUUAAGGUUUUUCUUAACGUAGCUG .........(((((((((((.......)))).))-)))))...(((((.(((((....((((....))))....)))))....(((((.((((.......)))).)))))...))))).. ( -35.70) >consensus AAAAAAAAAUCUUCGUUAGGUGAGGCUCCUGUAU_GGAGAUACGCUGCUGCCCAAAAAUGUCCAAAGACGCCAAUGGGUCAACAGAAAUCAUCGACAUAAGGUGAUUUUUAACGUAGCUG .........(((((((((((.......)))).)).)))))...(((((.(((((....((((....))))....)))))....((((((((((.......))))))))))...))))).. (-34.29 = -32.18 + -2.10)
| Location | 2,041,936 – 2,042,054 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -28.67 |
| Energy contribution | -26.90 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 2041936 118 + 4202352/40-160 UACGCUGCUGCCCAAAAACGUCCAAAGACGCCAAUGGGUCAACAGAAACCAUCGACAUAAGGUGGUUUUUAACGUAGCUGGCU--UCGUCCUAUGCCGCACAGUGCUAAAGCUCUACGAA ...(((((.(((((....((((....))))....)))))....((((((((((.......))))))))))...))))).(((.--.........)))....((.((....)).))..... ( -35.10) >Bclau.0 3492933 118 - 4303871/40-160 UACGCUGCUACCCAAAAAUGUCCAAAGACGCCAAUGGGUCAACAGAAACCAUCGACAUAAGGUGGUUUUUAAUGUAGCUGGCA--AUAGCCUAUGCCGCACAGUGCUAAAGCUCUACGAA ...(((((.(((((....((((....))))....)))))....((((((((((.......))))))))))...))))).((((--........))))....((.((....)).))..... ( -34.10) >Bsubt.0 1394917 120 + 4214630/40-160 UACGCUGCUGCCCAAAAAUGUCCAAAGACGCCAAUGGGUCAACAGAAAUCAUCGACAUAAGGUGAUUUUUAAUGCAGCUGGAUGCUUGUCCUAUGCCAUACAGUGCUAAAGCUCUACGAU ...(((((.(((((....((((....))))....)))))....((((((((((.......))))))))))...)))))((((.((((......((.....))......)))))))).... ( -32.80) >Blich.0 1419296 118 + 4222334/40-160 UACGCUGCUGCCCAAAAAUGUCCAAAGACGCCAAUGGGUCAACAGAAAUCAUCGAUCUAAGGUGAUUUUUAAUGCAGCUGCAU--GCUGCCUAUGCCAUACAGUGCUAAAGCUCUACGAU ...(((((.(((((....((((....))))....)))))....((((((((((.......))))))))))...))))).((((--(.....))))).....((.((....)).))..... ( -32.20) >Banth.0 1510394 119 + 5227293/40-160 AAUGCUACUGCCCAAAAAUGUCGAGAGACGCCAAUGGGUCAACAGGAAUGAUCGAAUUAAGGUUUUUCUUAACGUAGCUGGUAAUGUA-CCUAUGCUAUACAGUGCUAAAACUCGGCGAG ...(((((.(((((....((((....))))....)))))....(((((.((((.......)))).)))))...))))).((((...))-))..((((....(((......))).)))).. ( -29.20) >Bcere.0 1660585 119 + 5224283/40-160 AAUGCUACUGCCCAAAAAUGUCGAGAGACGCCAAUGGGUCAACAGGAAUGAUCGAAUUAAGGUUUUUCUUAACGUAGCUGGUAAUGUA-CCUAUGCUAUACAGUGCUAAAACUCGGCGAG ...(((((.(((((....((((....))))....)))))....(((((.((((.......)))).)))))...))))).((((...))-))..((((....(((......))).)))).. ( -29.20) >consensus UACGCUGCUGCCCAAAAAUGUCCAAAGACGCCAAUGGGUCAACAGAAAUCAUCGACAUAAGGUGAUUUUUAACGUAGCUGGAA__GUA_CCUAUGCCAUACAGUGCUAAAGCUCUACGAA ...(((((.(((((....((((....))))....)))))....((((((((((.......))))))))))...)))))((((............))))...(((......)))....... (-28.67 = -26.90 + -1.77)

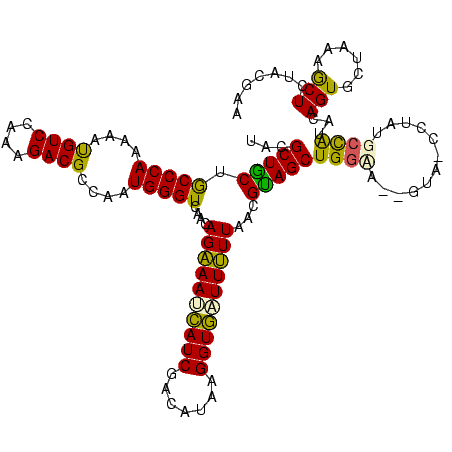
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:27:58 2006