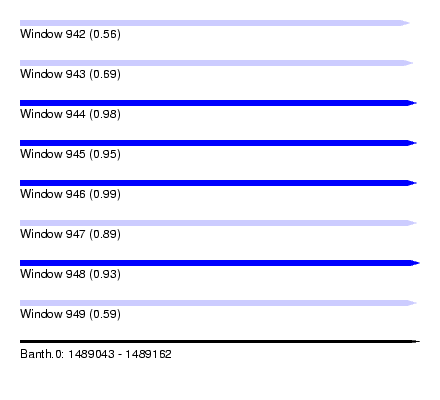
| Sequence ID | Banth.0 |
|---|---|
| Location | 1,489,043 – 1,489,162 |
| Length | 119 |
| Max. P | 0.989583 |

| Location | 1,489,043 – 1,489,159 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -39.75 |
| Consensus MFE | -25.38 |
| Energy contribution | -24.30 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
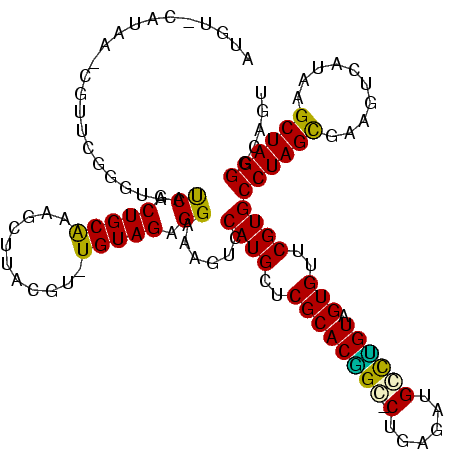
>Banth.0 1489043 116 + 5227293/0-120 AUGU-CAUAA-CGUUUGGGUAAUCGCUGCAACGCCAACGU-UGUAGAGGAAAGUCCAUGCUCGCACGGC-CUGAGAUGGCUGUAGUGUUCGUGCCUAGCCAAUUCAUAAGCUAGGGUAUU ....-(((((-(....(((((....((((((((....)))-))))).((.....)).)))))(((((((-(......)))))).))))).)))((((((..........))))))..... ( -36.40) >Bcere.0 1643226 116 + 5224283/0-120 AUGU-CAUAA-CGUUUGGGUAAUCGCUGCAACGCCAACGU-UGUAGAGGAAAGUCCAUGCUCGCACGGC-CUGAGAUGGCUGUAGUGUUCGUGCCUAGCCAAUUCAUAAGCUAGGGUAUU ....-(((((-(....(((((....((((((((....)))-))))).((.....)).)))))(((((((-(......)))))).))))).)))((((((..........))))))..... ( -36.40) >Bsubt.0 1883711 119 - 4214630/0-120 AUGUUCUUAA-CGUUCGGGUAAUCGCUGCAGAUCUUGAAUCUGUAGAGGAAAGUCCAUGCUCGCACGGUGCUGAGAUGCCCGUAGUGUUCGUGCCUAGCGAAGUCAUAAGCUAGGGCAGU ....(((((.-(((.((((((....((((((((.....)))))))).((.....)).)))))).)))....)))))(((((.((((.(((((.....))))).......))))))))).. ( -38.80) >Blich.0 1920987 118 - 4222334/0-120 AUGU-CUUAA-CGUUCGGGUAAUCGCUGCAGAUUUUACAUCUGUAGAGGAAAGUCCAUGCUCGCACGGUGCUGAGAUGCCCGUAGUGUUCGUGCCUAGCGAAGUCAUAAGCUAGGGCAGC ..((-((((.-(((.((((((....((((((((.....)))))))).((.....)).)))))).)))....))))))((((.((((.(((((.....))))).......))))))))... ( -39.20) >Bhalo.0 1843307 117 + 4202352/0-120 ACAU-CAUAACCGUUCAGGUAACCGCUGCAUCGCAUGCGA-UGUAGAGGAAAGUCCAUGCUCGCACAGG-CUGAGAUGCUUGUAGUGUUCGUGCCUAGCGAAGUCAUAAGCUAGGGUAGU ....-....(((.....)))..((.((((((((....)))-))))).))......((((..((((((((-(......)))))).)))..))))((((((..........))))))..... ( -41.90) >Bclau.0 2174225 116 - 4303871/0-120 ACAGAUACAUGUCUUCAGACAACCGCUGCGCAUUUUAUG--UGUAGAGGAAAGUCCAUGCUCGCACCGG-CUGAGAGGCCGGUAGUGUUCGUGCCUAGUUAAUCCAUAAGCUAGGGCAG- ...(((...((((....)))).((.((((((((...)))--))))).))...)))((((..((((((((-((....))))))).)))..))))(((((((........)))))))....- ( -45.80) >consensus AUGU_CAUAA_CGUUCGGGUAAUCGCUGCAAAGCUUACGU_UGUAGAGGAAAGUCCAUGCUCGCACGGC_CUGAGAUGCCUGUAGUGUUCGUGCCUAGCGAAGUCAUAAGCUAGGGCAGU ......................((.(((((...........))))).))......((((..((((((((.(......)))))).)))..))))((((((..........))))))..... (-25.38 = -24.30 + -1.08)

| Location | 1,489,043 – 1,489,160 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -41.37 |
| Consensus MFE | -25.13 |
| Energy contribution | -24.20 |
| Covariance contribution | -0.93 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 1489043 117 + 5227293/40-160 -UGUAGAGGAAAGUCCAUGCUCGCACGGC-CUGAGAUGGCUGUAGUGUUCGUGCCUAGCCAAUUCAUAAGCUAGGGUAUUCUGGCUGUAAGG-CUGGUUUAACGGCAGGGAAAAAACCUA -.....(((....(((.(((.((((((((-(......)))))).)))..(((..((((((........(((((((....)))))))....))-))))....)))))).))).....))). ( -39.70) >Bcere.0 1643226 117 + 5224283/40-160 -UGUAGAGGAAAGUCCAUGCUCGCACGGC-CUGAGAUGGCUGUAGUGUUCGUGCCUAGCCAAUUCAUAAGCUAGGGUAUUCUGGCUGUAAGG-CUGGUUUAACGGCAGGGAAAAAACCUA -.....(((....(((.(((.((((((((-(......)))))).)))..(((..((((((........(((((((....)))))))....))-))))....)))))).))).....))). ( -39.70) >Bsubt.0 1883711 111 - 4214630/40-160 CUGUAGAGGAAAGUCCAUGCUCGCACGGUGCUGAGAUGCCCGUAGUGUUCGUGCCUAGCGAAGUCAUAAGCUAGGGCAGUCUU--UA-GAGG-CUG-----ACGGCAGGAAAAAAGCCUA ......(((....(((.(((.(((((((.((......)))))).)))..(((.((((((..........)))))).((((((.--..-.)))-)))-----)))))))))......))). ( -38.30) >Blich.0 1920987 113 - 4222334/40-160 CUGUAGAGGAAAGUCCAUGCUCGCACGGUGCUGAGAUGCCCGUAGUGUUCGUGCCUAGCGAAGUCAUAAGCUAGGGCAGCCUUGCAG-AAGG-CUG-----ACGGCAGGAAAAAAGCCUA ......(((....(((.(((.(((((((.((......)))))).)))..(((.((((((..........)))))).(((((((....-))))-)))-----)))))))))......))). ( -42.10) >Bhalo.0 1843307 118 + 4202352/40-160 -UGUAGAGGAAAGUCCAUGCUCGCACAGG-CUGAGAUGCUUGUAGUGUUCGUGCCUAGCGAAGUCAUAAGCUAGGGUAGUCUGGUUCUUAGAACUGGGCUAACGGCAAGGUAAGCACCUA -.....(((.....((.(((.((((((((-(......)))))).)))..(((.((((((..........)))))).(((((..((((...))))..))))))))))).))......))). ( -41.00) >Bclau.0 2174225 110 - 4303871/40-160 -UGUAGAGGAAAGUCCAUGCUCGCACCGG-CUGAGAGGCCGGUAGUGUUCGUGCCUAGUUAAUCCAUAAGCUAGGGCAG-CUGCUGUGCAG-------CUGACGGCAGGAGAGAUGCCUA -.....(((....(((.(((.((((((((-((....))))))).)))..(((.(((((((........))))))).(((-((((...))))-------))))))))))))......))). ( -47.40) >consensus _UGUAGAGGAAAGUCCAUGCUCGCACGGC_CUGAGAUGCCUGUAGUGUUCGUGCCUAGCGAAGUCAUAAGCUAGGGCAGUCUGGCUGUAAGG_CUG___UAACGGCAGGAAAAAAACCUA ......(((....(((.(((.((((((((.(......)))))).)))..(((.((((((..........))))))....((((.....)))).........)))))))))......))). (-25.13 = -24.20 + -0.93)

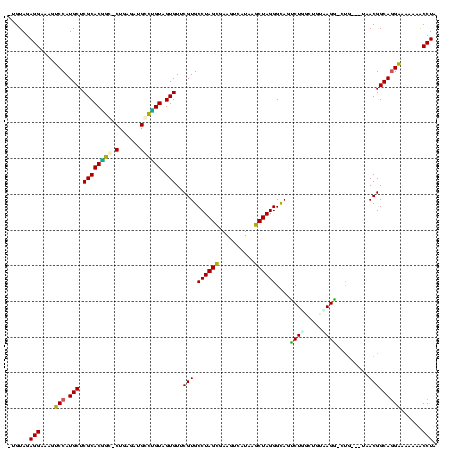
| Location | 1,489,043 – 1,489,161 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -38.19 |
| Consensus MFE | -26.97 |
| Energy contribution | -25.61 |
| Covariance contribution | -1.36 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 1489043 118 + 5227293/80-200 UGUAGUGUUCGUGCCUAGCCAAUUCAUAAGCUAGGGUAUUCUGGCUGUAAGG-CUGGUUUAACGGCAGGGAAAAAACCUAAGUCCUUU-CGGAUAUGGUUUGACUACCUUUAAAGUGCCA .(((((.(((.((((.(((((.......(((((((....)))))))......-.)))))....)))).)))..(((((...((((...-.))))..))))).)))))............. ( -34.74) >Bcere.0 1643226 118 + 5224283/80-200 UGUAGUGUUCGUGCCUAGCCAAUUCAUAAGCUAGGGUAUUCUGGCUGUAAGG-CUGGUUUAACGGCAGGGAAAAAACCUAAGUCCUUU-CGGAUAUGGUUUGACUACCUUUAAAGUGCCA .(((((.(((.((((.(((((.......(((((((....)))))))......-.)))))....)))).)))..(((((...((((...-.))))..))))).)))))............. ( -34.74) >Bsubt.0 1883711 108 - 4214630/80-200 CGUAGUGUUCGUGCCUAGCGAAGUCAUAAGCUAGGGCAGUCUU--UA-GAGG-CUG-----ACGGCAGGAAAAAAGCCUACGUCUUC---GGAUAUGGCUGAGUAUCCUUGAAAGUGCCA ..........((.((((((..........)))))).((((((.--..-.)))-)))-----))(((((((.(..((((...(((...---.)))..))))...).))).......)))). ( -33.81) >Blich.0 1920987 113 - 4222334/80-200 CGUAGUGUUCGUGCCUAGCGAAGUCAUAAGCUAGGGCAGCCUUGCAG-AAGG-CUG-----ACGGCAGGAAAAAAGCCUACGUCCUUUAUGGAUAUGGCUGAGUAUCCUUGAAAGUGCCA ..........((.((((((..........)))))).(((((((....-))))-)))-----))(((((((.(..((((...((((.....))))..))))...).))).......)))). ( -38.11) >Bhalo.0 1843307 120 + 4202352/80-200 UGUAGUGUUCGUGCCUAGCGAAGUCAUAAGCUAGGGUAGUCUGGUUCUUAGAACUGGGCUAACGGCAAGGUAAGCACCUACGUUCAUCAUGAAUAUGGUGUGAUGCCUUUGAAAGUGCCA ..........((.((((((..........)))))).(((((..((((...))))..)))))))(((((((((.(((((...((((.....))))..)))))..))))).......)))). ( -41.71) >Bclau.0 2174225 111 - 4303871/80-200 GGUAGUGUUCGUGCCUAGUUAAUCCAUAAGCUAGGGCAG-CUGCUGUGCAG-------CUGACGGCAGGAGAGAUGCCUACGUCCAA-AUGGAUAAGGCAUGCACUCCUUGAAAGUGCCA ((((.(((.(((.(((((((........))))))).(((-((((...))))-------))))))))(((((.(((((((..((((..-..)))).)))))).).)))))....).)))). ( -46.00) >consensus UGUAGUGUUCGUGCCUAGCGAAGUCAUAAGCUAGGGCAGUCUGGCUGUAAGG_CUG___UAACGGCAGGAAAAAAACCUACGUCCUUU_UGGAUAUGGCUUGAUAUCCUUGAAAGUGCCA .............((((((..........))))))............................((((((((..(((((...((((.....))))..)))))...)))).......)))). (-26.97 = -25.61 + -1.36)

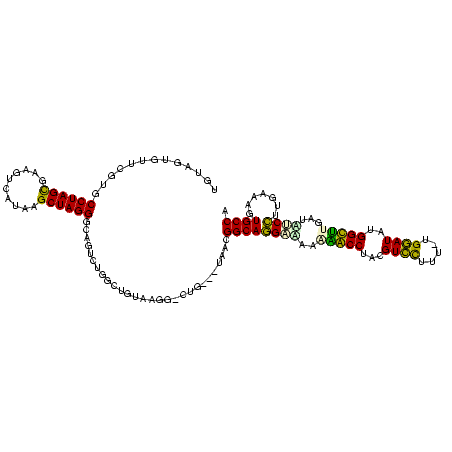
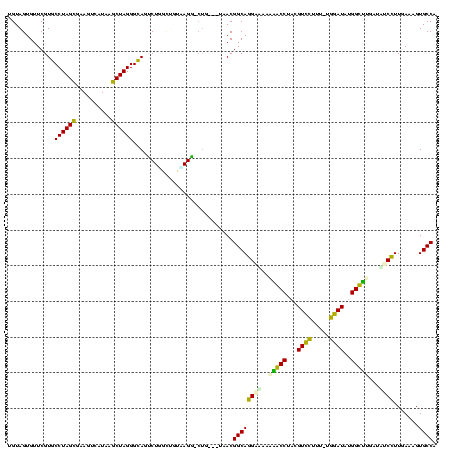
| Location | 1,489,043 – 1,489,161 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -20.06 |
| Energy contribution | -19.18 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 1489043 118 + 5227293/80-200 UGGCACUUUAAAGGUAGUCAAACCAUAUCCG-AAAGGACUUAGGUUUUUUCCCUGCCGUUAAACCAG-CCUUACAGCCAGAAUACCCUAGCUUAUGAAUUGGCUAGGCACGAACACUACA .(((..(((((.(((((..((((((..(((.-...)))..).))))).....))))).)))))...)-))...............(((((((........)))))))............. ( -32.00) >Bcere.0 1643226 118 + 5224283/80-200 UGGCACUUUAAAGGUAGUCAAACCAUAUCCG-AAAGGACUUAGGUUUUUUCCCUGCCGUUAAACCAG-CCUUACAGCCAGAAUACCCUAGCUUAUGAAUUGGCUAGGCACGAACACUACA .(((..(((((.(((((..((((((..(((.-...)))..).))))).....))))).)))))...)-))...............(((((((........)))))))............. ( -32.00) >Bsubt.0 1883711 108 - 4214630/80-200 UGGCACUUUCAAGGAUACUCAGCCAUAUCC---GAAGACGUAGGCUUUUUUCCUGCCGU-----CAG-CCUC-UA--AAGACUGCCCUAGCUUAUGACUUCGCUAGGCACGAACACUACG .(((........(((((........)))))---...((((((((.......)))).)))-----).)-))..-..--.....((((.((((..........))))))))........... ( -27.20) >Blich.0 1920987 113 - 4222334/80-200 UGGCACUUUCAAGGAUACUCAGCCAUAUCCAUAAAGGACGUAGGCUUUUUUCCUGCCGU-----CAG-CCUU-CUGCAAGGCUGCCCUAGCUUAUGACUUCGCUAGGCACGAACACUACG .((((.......(((.....(((((..(((.....)))..).))))....)))))))(.-----(((-((((-....))))))))((((((..........))))))............. ( -33.01) >Bhalo.0 1843307 120 + 4202352/80-200 UGGCACUUUCAAAGGCAUCACACCAUAUUCAUGAUGAACGUAGGUGCUUACCUUGCCGUUAGCCCAGUUCUAAGAACCAGACUACCCUAGCUUAUGACUUCGCUAGGCACGAACACUACA .(((((((.......(((((...........))))).....)))))))...(.((((..((((..((((.((((....((......))..)))).))))..)))))))).)......... ( -25.10) >Bclau.0 2174225 111 - 4303871/80-200 UGGCACUUUCAAGGAGUGCAUGCCUUAUCCAU-UUGGACGUAGGCAUCUCUCCUGCCGUCAG-------CUGCACAGCAG-CUGCCCUAGCUUAUGGAUUAACUAGGCACGAACACUACC .((((.......((((.(.((((((..(((..-..)))...))))))).))))))))(.(((-------((((...))))-))))(((((.(((.....))))))))............. ( -38.81) >consensus UGGCACUUUCAAGGAAAUCAAACCAUAUCCA_AAAGGACGUAGGCUUUUUCCCUGCCGUUA___CAG_CCUUACAGCCAGACUACCCUAGCUUAUGAAUUCGCUAGGCACGAACACUACA .((((.......(((.....(((((..(((.....)))..).))))....)))))))............................((((((..........))))))............. (-20.06 = -19.18 + -0.89)

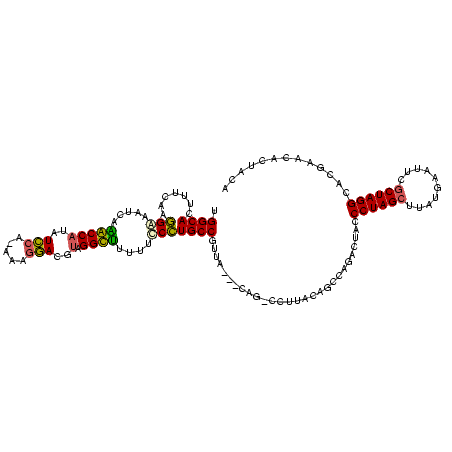
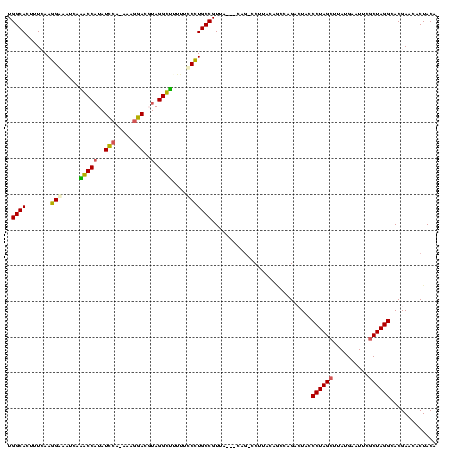
| Location | 1,489,043 – 1,489,161 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.85 |
| Mean single sequence MFE | -37.35 |
| Consensus MFE | -31.38 |
| Energy contribution | -30.41 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
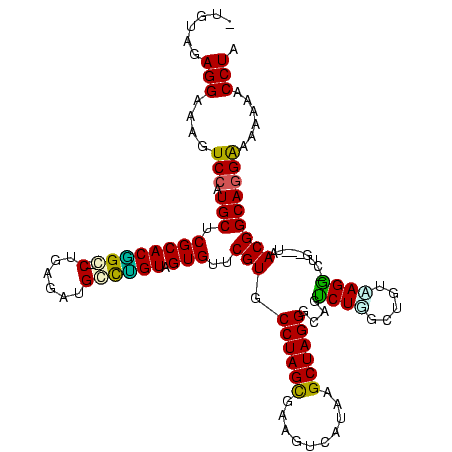
>Banth.0 1489043 118 + 5227293/120-240 CUGGCUGUAAGG-CUGGUUUAACGGCAGGGAAAAAACCUAAGUCCUUU-CGGAUAUGGUUUGACUACCUUUAAAGUGCCACAGUGACGAAGUCCUUGAAGAAAUGAUAGGAGUGGAACGA .((((......(-(((......))))(((....(((((...((((...-.))))..))))).....))).......))))..((..((...((((...(....)...)))).))..)).. ( -30.00) >Bcere.0 1643226 118 + 5224283/120-240 CUGGCUGUAAGG-CUGGUUUAACGGCAGGGAAAAAACCUAAGUCCUUU-CGGAUAUGGUUUGACUACCUUUAAAGUGCCACAGUGACGAAGUCCUUGAAGAAAUGAUAGGAGUGGAACGA .((((......(-(((......))))(((....(((((...((((...-.))))..))))).....))).......))))..((..((...((((...(....)...)))).))..)).. ( -30.00) >Bsubt.0 1883711 108 - 4214630/120-240 CUU--UA-GAGG-CUG-----ACGGCAGGAAAAAAGCCUACGUCUUC---GGAUAUGGCUGAGUAUCCUUGAAAGUGCCACAGUGACGAAGUCUCACUAGAAAUGGUGAGAGUGGAACGC ...--..-..((-(..-----.....((((.(..((((...(((...---.)))..))))...).)))).......)))...((..((...((((((((....)))))))).))..)).. ( -35.04) >Blich.0 1920987 113 - 4222334/120-240 CUUGCAG-AAGG-CUG-----ACGGCAGGAAAAAAGCCUACGUCCUUUAUGGAUAUGGCUGAGUAUCCUUGAAAGUGCCACAGUGACGAAGUCUCACUAGAAAUGGUGAGAGUGGAACGC ...((..-..((-(.(-----(((..(((.......))).))))((((..((((((......))))))...)))).)))...((..((...((((((((....)))))))).))..)))) ( -36.90) >Bhalo.0 1843307 120 + 4202352/120-240 CUGGUUCUUAGAACUGGGCUAACGGCAAGGUAAGCACCUACGUUCAUCAUGAAUAUGGUGUGAUGCCUUUGAAAGUGCCACAGUGACGUAGUCCGUUUGGAAACAGACGGAGUGGAACGA ...((((.....(((((((.......((((((.(((((...((((.....))))..)))))..)))))).......))).))))....((.((((((((....)))))))).)))))).. ( -46.04) >Bclau.0 2174225 112 - 4303871/120-240 CUGCUGUGCAG-------CUGACGGCAGGAGAGAUGCCUACGUCCAA-AUGGAUAAGGCAUGCACUCCUUGAAAGUGCCACAGUGACGUAGUCUGCUUGGAAACGGGCAGAGUGGAACGC ..((((((..(-------((..((..(((((.(((((((..((((..-..)))).)))))).).)))))))..)))..))))))....((.((((((((....)))))))).))...... ( -46.10) >consensus CUGGCUGUAAGG_CUG___UAACGGCAGGAAAAAAACCUACGUCCUUU_UGGAUAUGGCUUGAUAUCCUUGAAAGUGCCACAGUGACGAAGUCCCUCUAGAAAUGGUAAGAGUGGAACGA .......................((((((((..(((((...((((.....))))..)))))...)))).......))))...((..((...((((((((....)))))))).))..)).. (-31.38 = -30.41 + -0.96)

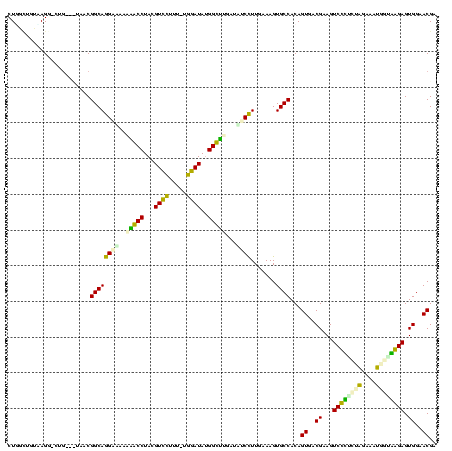
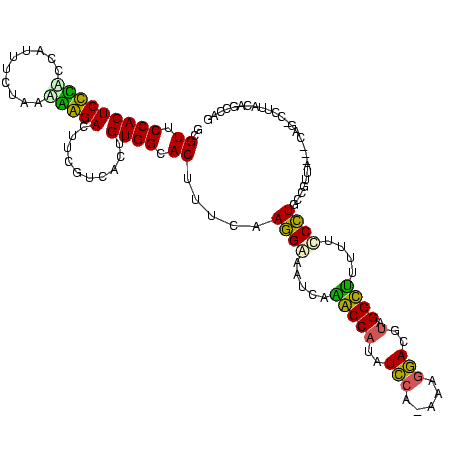
| Location | 1,489,043 – 1,489,161 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.85 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -19.26 |
| Energy contribution | -17.10 |
| Covariance contribution | -2.16 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 1489043 118 + 5227293/120-240 UCGUUCCACUCCUAUCAUUUCUUCAAGGACUUCGUCACUGUGGCACUUUAAAGGUAGUCAAACCAUAUCCG-AAAGGACUUAGGUUUUUUCCCUGCCGUUAAACCAG-CCUUACAGCCAG .........((((............))))........(((((((..(((((.(((((..((((((..(((.-...)))..).))))).....))))).)))))...)-))..)))).... ( -26.30) >Bcere.0 1643226 118 + 5224283/120-240 UCGUUCCACUCCUAUCAUUUCUUCAAGGACUUCGUCACUGUGGCACUUUAAAGGUAGUCAAACCAUAUCCG-AAAGGACUUAGGUUUUUUCCCUGCCGUUAAACCAG-CCUUACAGCCAG .........((((............))))........(((((((..(((((.(((((..((((((..(((.-...)))..).))))).....))))).)))))...)-))..)))).... ( -26.30) >Bsubt.0 1883711 108 - 4214630/120-240 GCGUUCCACUCUCACCAUUUCUAGUGAGACUUCGUCACUGUGGCACUUUCAAGGAUACUCAGCCAUAUCC---GAAGACGUAGGCUUUUUUCCUGCCGU-----CAG-CCUC-UA--AAG (((......((((((........))))))...)))....(.(((........(((((........)))))---...((((((((.......)))).)))-----).)-)).)-..--... ( -25.80) >Blich.0 1920987 113 - 4222334/120-240 GCGUUCCACUCUCACCAUUUCUAGUGAGACUUCGUCACUGUGGCACUUUCAAGGAUACUCAGCCAUAUCCAUAAAGGACGUAGGCUUUUUUCCUGCCGU-----CAG-CCUU-CUGCAAG ((.......((((((........))))))........((((((((.......(((.....(((((..(((.....)))..).))))....)))))))).-----)))-....-..))... ( -28.51) >Bhalo.0 1843307 120 + 4202352/120-240 UCGUUCCACUCCGUCUGUUUCCAAACGGACUACGUCACUGUGGCACUUUCAAAGGCAUCACACCAUAUUCAUGAUGAACGUAGGUGCUUACCUUGCCGUUAGCCCAGUUCUAAGAACCAG ..((((......(((((((....)))))))......((((.(((.......((((.....(((((..(((.....)))..).))))....)))).......))))))).....))))... ( -29.14) >Bclau.0 2174225 112 - 4303871/120-240 GCGUUCCACUCUGCCCGUUUCCAAGCAGACUACGUCACUGUGGCACUUUCAAGGAGUGCAUGCCUUAUCCAU-UUGGACGUAGGCAUCUCUCCUGCCGUCAG-------CUGCACAGCAG ((.......(((((..........)))))....((((((((((((.......((((.(.((((((..(((..-..)))...))))))).))))))))).)))-------.)).)).)).. ( -35.91) >consensus GCGUUCCACUCCCACCAUUUCUAAAAAGACUUCGUCACUGUGGCACUUUCAAGGAAAUCAAACCAUAUCCA_AAAGGACGUAGGCUUUUUCCCUGCCGUUA___CAG_CCUUACAGCCAG ..((.(((((((((..........)))))..........)))).)).....((((.....(((((..(((.....)))..).))))....)))).......................... (-19.26 = -17.10 + -2.16)

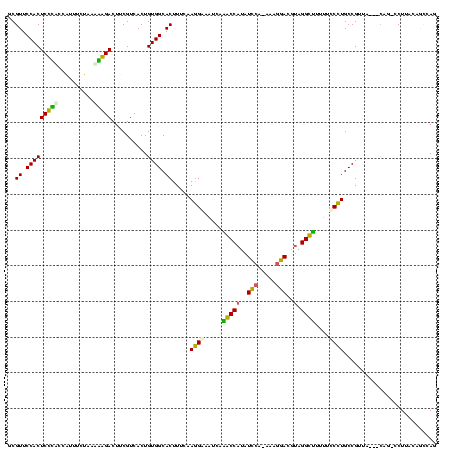
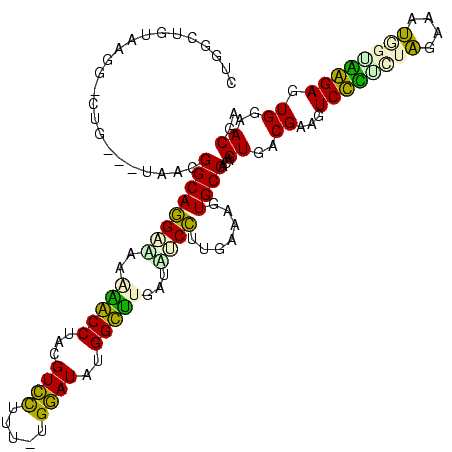
| Location | 1,489,043 – 1,489,162 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.10 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -25.48 |
| Energy contribution | -25.32 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 1489043 119 + 5227293/200-320 CAGUGACGAAGUCCUUGAAGAAAUGAUAGGAGUGGAACGAGGUAAACCCCACGAGCGAGAAACCCAAAUAAUGGUAGGGGAAUCUUUUCCAA-GGAAAUGAACGAUGGGAAAGGACAGGU ..((..((...((((...(....)...)))).))..))........((((.(......)..(((........))).))))..(((((((((.-(........)..)))))))))...... ( -24.70) >Bcere.0 1643226 119 + 5224283/200-320 CAGUGACGAAGUCCUUGAAGAAAUGAUAGGAGUGGAACGAGGUAAACCCCACGAGCGAGAAACCCAAAUAAUGGUAGGGGAAUCUUUUCCAA-GGAAAUGAACGAUGGGAAAGGACAGGU ..((..((...((((...(....)...)))).))..))........((((.(......)..(((........))).))))..(((((((((.-(........)..)))))))))...... ( -24.70) >Bsubt.0 1883711 118 - 4214630/200-320 CAGUGACGAAGUCUCACUAGAAAUGGUGAGAGUGGAACGCGGUAAACCCCUCGAGCGAGAAACCCAAAUUUUGGUAGGGGAACCUUCUUAAC-GGAAUUCAACGG-AGAGAAGGACAGAA ..(((....(.((((((((....)))))))).)....)))......(((((..(.(((((........))))).))))))..(((((((..(-(........)).-.)))))))...... ( -39.00) >Blich.0 1920987 118 - 4222334/200-320 CAGUGACGAAGUCUCACUAGAAAUGGUGAGAGUGGAACGCGGUAAACCCCUCGAGCGAGAAACCCAAAUUUUGGUAGGGGAACCUUCUGGAC-GGAAAUGAACGG-AAGGAAGGACAGAG ..(((....(.((((((((....)))))))).)....)))......(((((..(.(((((........))))).))))))..((((((...(-(........)).-..))))))...... ( -37.30) >Bhalo.0 1843307 118 + 4202352/200-320 CAGUGACGUAGUCCGUUUGGAAACAGACGGAGUGGAACGAGGUAAACCCCUCGAGCGAGAAACCCAAAAAUUGGUAGGGGAACCUUCUUGAA-GGAAAUGAACGGAGA-GAAGGACGGAA ......((((.((((((((....)))))))).)....(((((......))))).))).....(((...........)))...(((((((...-(........)...))-)))))...... ( -38.00) >Bclau.0 2174225 120 - 4303871/200-320 CAGUGACGUAGUCUGCUUGGAAACGGGCAGAGUGGAACGCGGUAAACCCCUCGAGCGAGAAACCCAAACAAUGGUAGGGGCAUUUUUCAGUCUGAAAAUGAACGGAACUGAAAAAACGAA ..(((...((.((((((((....)))))))).))...)))......((((((......)).(((........))).))))..((((((((((((........))).)))))))))..... ( -37.30) >consensus CAGUGACGAAGUCCCUCUAGAAAUGGUAAGAGUGGAACGAGGUAAACCCCUCGAGCGAGAAACCCAAAUAAUGGUAGGGGAACCUUCUCGAA_GGAAAUGAACGG_AGGGAAGGACAGAA ..((..((...((((((((....)))))))).))..))........((((((......)).(((........))).))))..(((((((....(........)....)))))))...... (-25.48 = -25.32 + -0.16)
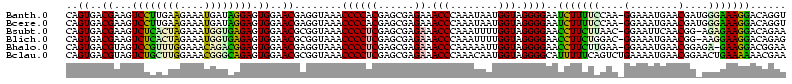
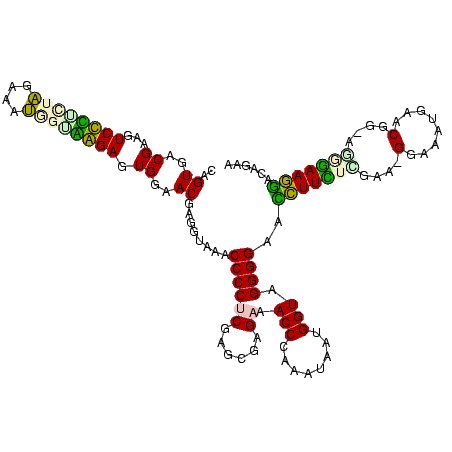
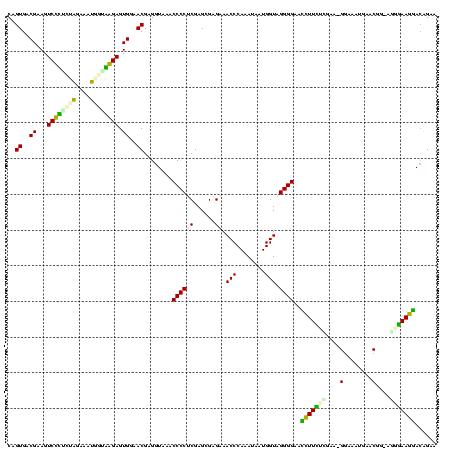
| Location | 1,489,043 – 1,489,161 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.73 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -19.39 |
| Energy contribution | -18.40 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 1489043 118 + 5227293/240-360 GGUAAACCCCACGAGCGAGAAACCCAAAUAAUGGUAGGGGAAUCUUUUCCAA-GGAAAUGAACGAUGGGAAAGGACAGGUUGUAUAA-CCUGUAGAUAGAUGAUUGCCACCGGAGUACGA (((((.((((.(......)..(((........))).))))..(((((((((.-(........)..)))))))))(((((((....))-)))))..........)))))..((.....)). ( -31.20) >Bcere.0 1643226 118 + 5224283/240-360 GGUAAACCCCACGAGCGAGAAACCCAAAUAAUGGUAGGGGAAUCUUUUCCAA-GGAAAUGAACGAUGGGAAAGGACAGGUUGUAUAA-CCUGUAGAUAGAUGAUUGCCACCGGAGUACGA (((((.((((.(......)..(((........))).))))..(((((((((.-(........)..)))))))))(((((((....))-)))))..........)))))..((.....)). ( -31.20) >Bsubt.0 1883711 115 - 4214630/240-360 GGUAAACCCCUCGAGCGAGAAACCCAAAUUUUGGUAGGGGAACCUUCUUAAC-GGAAUUCAACGG-AGAGAAGGACAGAAUGCUU---UCUGUAGAUAGAUGAUUGCCGCCUGAGUACGA (((((.(((((..(.(((((........))))).))))))..(((((((..(-(........)).-.)))))))((((((....)---)))))..........)))))............ ( -33.70) >Blich.0 1920987 115 - 4222334/240-360 GGUAAACCCCUCGAGCGAGAAACCCAAAUUUUGGUAGGGGAACCUUCUGGAC-GGAAAUGAACGG-AAGGAAGGACAGAGUUGAC---UCUGUAGAUAGAUGAUUACCACCCGAGUACGA (((((.(((((..(.(((((........))))).))))))..((((((...(-(........)).-..))))))((((((....)---)))))..........)))))............ ( -34.00) >Bhalo.0 1843307 118 + 4202352/240-360 GGUAAACCCCUCGAGCGAGAAACCCAAAAAUUGGUAGGGGAACCUUCUUGAA-GGAAAUGAACGGAGA-GAAGGACGGAAGUUUUACUUCUGUAGACAGAUGGUUACCACCGGAGUACGA (((((.((((((......)).(((........))).))))..(((((((...-(........)...))-)))))(((((((.....)))))))..........)))))..((.....)). ( -32.60) >Bclau.0 2174225 116 - 4303871/240-360 GGUAAACCCCUCGAGCGAGAAACCCAAACAAUGGUAGGGGCAUUUUUCAGUCUGAAAAUGAACGGAACUGAAAAAACGAAG---CAAUUC-GUAGAUAGAUGGUUGUCGCCGGAGUACGA .(((..((.(((....)))..(((........)))...(((.((((((((((((........))).)))))))))(((((.---...)))-)).(((((....))))))))))..))).. ( -30.60) >consensus GGUAAACCCCUCGAGCGAGAAACCCAAAUAAUGGUAGGGGAACCUUCUCGAA_GGAAAUGAACGG_AGGGAAGGACAGAAUGUAUAA_UCUGUAGAUAGAUGAUUGCCACCGGAGUACGA (((((.((((((......)).(((........))).))))..(((((((....(........)....)))))))(((((.........)))))..........)))))............ (-19.39 = -18.40 + -0.99)

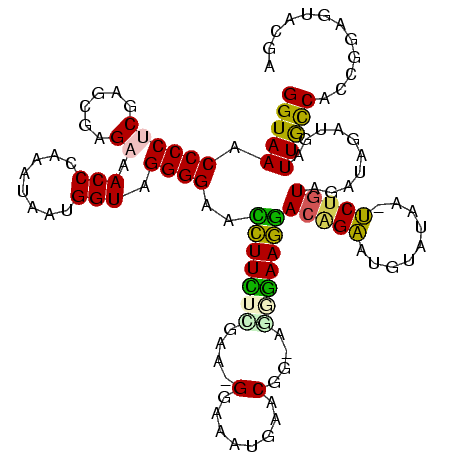
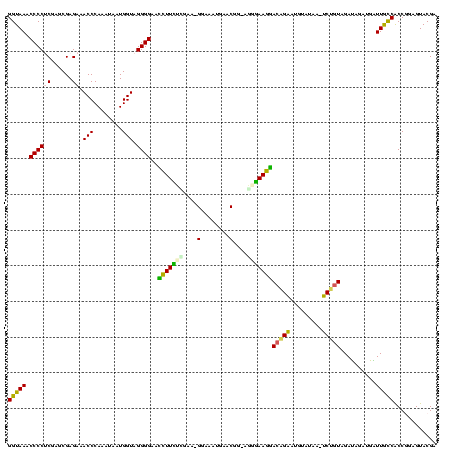
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:27:56 2006