| Sequence ID | Bsubt.0 |
|---|---|
| Location | 1,783,825 – 1,783,935 |
| Length | 110 |
| Max. P | 0.912041 |

| Location | 1,783,825 – 1,783,935 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.81 |
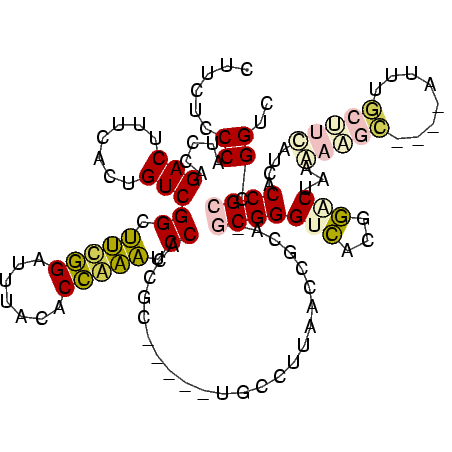
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -15.67 |
| Energy contribution | -15.40 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
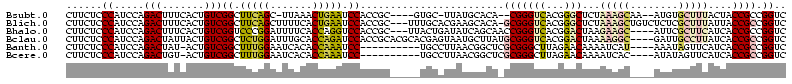
>Bsubt.0 1783825 110 - 4214630/23-143 CUUCUCCCAUCCAGACUUUCACUGUCGGCUUCAGC-UUAAACUGAAUCCACCGC----GUGC-UUAUGCACA--CGGGUCACGGGCUCUAAAGCAA--AUGUGCUUUACUACCGCCGGUC ......((.....(((..........((.(((((.-.....))))).)).(((.----((((-....)))).--)))))).(((....(((((((.--...)))))))...)))..)).. ( -33.80) >Blich.0 1813934 116 - 4222334/23-143 CUUCUCCCAUCCAGACUUUCACUGUCGGCUUCAGCUUUUCACUGAAUCCACCGC---UUUGCACGAAGCACA-GCGGGUCACGGGCUCUAAAGCUGUCUCUCGCUUUAUUACCGCCGGUC ......((.....(((..........((.(((((.......))))).)).((((---(.(((.....))).)-))))))).(((....((((((........))))))...)))..)).. ( -33.00) >Bhalo.0 1634620 113 + 4202352/23-143 CUUCUCCCAUCCAGACUUUCACUGUCGGUCCCGGAUUUUCACCAGGUCCACCGC---UUACUGAUAUCAGCAACCGGGUCACGGACUAAGAAGC----AUUCGCUUCAUCACCGCCGGUC ..........((.(((.......)))))..((((..........(((((.(((.---((.(((....))).)).))).....)))))..(((((----....))))).......)))).. ( -29.20) >Bclau.0 1930366 116 + 4303871/23-143 CUUCUCCCAUCCAGACUAUUACUGUCGGCUCUGGAUUUGCACCAGAUCCACCGCACGCACGAGUAAUGCUUAUGCGGGUCACGGACUAAAAGGC----GAUUGCCUUAUCACCGCCGGUC ......((.....((((.....((.(((...((((((((...)))))))))))))((((..((.....))..)))))))).(((.....(((((----....)))))....)))..)).. ( -34.10) >Banth.0 1422570 105 + 5227293/23-143 CUUCUCCCAUCCAGACUAU-ACUGUCGGCUUUGGAAUCACACCAAAUCC----------UGCCUUAACGGCUCGCGGGCUUAGAACAAAAUCAU----AAAUAGUUCAUCACCGCCGGUC .............(((...-...)))((((((((.......)))))...----------.))).....((((.((((.....((((........----.....))))....)))).)))) ( -22.42) >Bcere.0 1576748 105 + 5224283/23-143 CUUCUCCCAUCCAGACUGU-ACUGUCGGCUUUGGAAUCACACCAAAUCC----------UGCCUUAACGGCUCGCGGGCUUAGAACAAAAUCAC----AUAUAGUUCAUCACCGCCGGUC .........(((((((((.-.....))).)))))).....(((......----------.(((.....)))..((((.....((((........----.....))))....)))).))). ( -23.42) >consensus CUUCUCCCAUCCAGACUUUCACUGUCGGCUUCGGAUUUACACCAAAUCCACCGC_____UGCCUUAACCGCA_GCGGGUCACGGACUAAAAAGC____AUUUGCUUCAUCACCGCCGGUC ......((.....(((.......)))((.(((((.......))))).))........................(((((((...)))...(((((........)))))....)))).)).. (-15.67 = -15.40 + -0.27)
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:27:50 2006