
| Sequence ID | Blich.0 |
|---|---|
| Location | 4,187,073 – 4,187,193 |
| Length | 120 |
| Max. P | 0.998899 |

| Location | 4,187,073 – 4,187,178 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.21 |
| Mean single sequence MFE | -37.61 |
| Consensus MFE | -31.20 |
| Energy contribution | -30.20 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
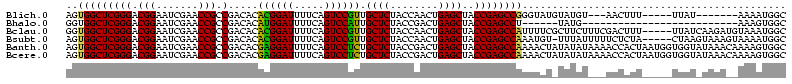
>Blich.0 4187073 105 + 4222334/0-120 AGUGGCUCGGGACGGAAUCGAACCGCCGACACACGGAUUUUCAGUCCGUUGCUCUACCAACUGAGCUACCGAGCCGGGUAUGUAUGU---AACUUU-----UUAU-------AAAAUGGC ..((((((((((((((...(((...(((.....)))...)))..))))))((((........))))..))))))))...........---......-----....-------........ ( -34.70) >Bhalo.0 4163644 88 + 4202352/0-120 GGUGGCUCGGGACGGAAUCGAACCGCCGACACAUGGAUUUUCAGUCCAUUGCUCUACCGACUGAGCUACCGAGCCU------UAUG--------------------------AAAGUGGC ((((((((((..(((..(((......)))((.((((((.....)))))))).....))).))))))))))..(((.------....--------------------------.....))) ( -32.40) >Bclau.0 4281557 115 + 4303871/0-120 GGUGGCUCGGGACGGAAUCGAACCGCCGACACACGGAUUUUCAGUCCGUUGCUCUACCAACUGAGCUACCGAGCCAUUUUCGCUUCUUUCGACUUU-----UUAUCAAGAUGUAAAUGGC ((((((((((((((((...(((...(((.....)))...)))..))))))((((........))))..))))))))))...(((.....((.(((.-----.....))).)).....))) ( -38.40) >Bsubt.0 4153807 114 + 4214630/0-120 AGUGGCUCGGGACGGAAUCGAACCGCCGACACACGGAUUUUCAGUCCGUUGCUCUACCAACUGAGCUACCGAGCCAAAUGU-UUUAUUUUUCUCUA-----CUAAGUAAAGUAAAAUGGC ..((((((((((((((...(((...(((.....)))...)))..))))))((((........))))..))))))))...((-((((((((.((...-----...)).))))))))))... ( -40.80) >Banth.0 5206672 120 + 5227293/0-120 AGUGGCUCGGGACGGAAUCGAACCGCCGACACGAGGAUUUUCAGUCCUCUGCUCUACCGACUGAGCUACCGAGCCAAAACUAUAUAUAAAACCACUAAUGGUGGUAUAAACAAAAGUGGC .(((((((((..(((..(((......)))...((((((.....)))))).......))).)))))))))...((((........((((..((((....))))..))))........)))) ( -39.69) >Bcere.0 5203671 120 + 5224283/0-120 AGUGGCUCGGGACGGAAUCGAACCGCCGACACGAGGAUUUUCAGUCCUCUGCUCUACCGACUGAGCUACCGAGCCAAAACUAUAUAUAAAACCACUAAUGGUGGUAUAAACAAAAGUGGC .(((((((((..(((..(((......)))...((((((.....)))))).......))).)))))))))...((((........((((..((((....))))..))))........)))) ( -39.69) >consensus AGUGGCUCGGGACGGAAUCGAACCGCCGACACACGGAUUUUCAGUCCGUUGCUCUACCAACUGAGCUACCGAGCCAAAUCU_UAUAU___ACCAUU_____UUAU_UAAA__AAAAUGGC ..(((((((((.(((.......))).).....((((((.....)))))).((((........))))..))))))))............................................ (-31.20 = -30.20 + -1.00)
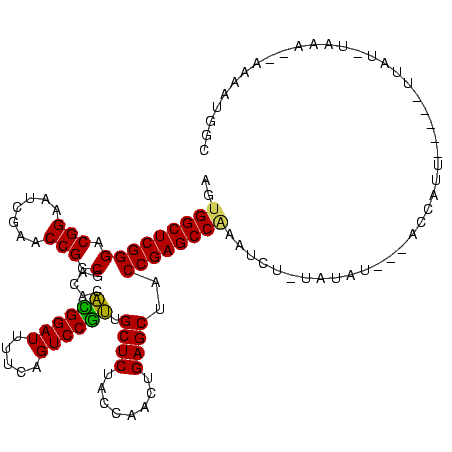
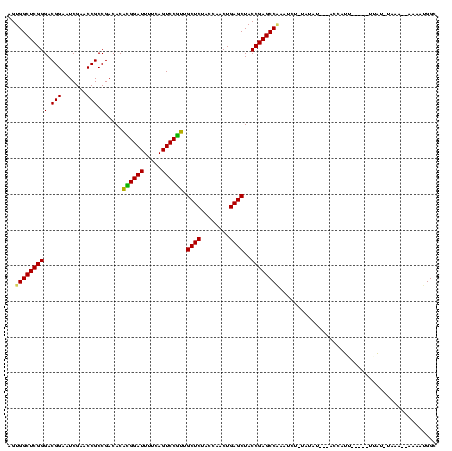
| Location | 4,187,073 – 4,187,178 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.21 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -33.34 |
| Energy contribution | -32.28 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
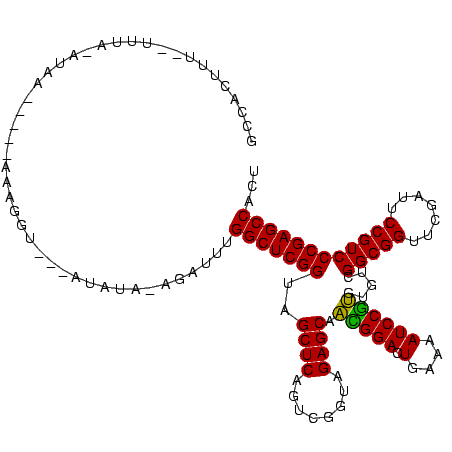
>Blich.0 4187073 105 + 4222334/0-120 GCCAUUUU-------AUAA-----AAAGUU---ACAUACAUACCCGGCUCGGUAGCUCAGUUGGUAGAGCAACGGACUGAAAAUCCGUGUGUCGGCGGUUCGAUUCCGUCCCGAGCCACU ........-------....-----......---............(((((((..((((........)))).(((((.(....)))))).....(((((.......))))))))))))... ( -32.30) >Bhalo.0 4163644 88 + 4202352/0-120 GCCACUUU--------------------------CAUA------AGGCUCGGUAGCUCAGUCGGUAGAGCAAUGGACUGAAAAUCCAUGUGUCGGCGGUUCGAUUCCGUCCCGAGCCACC ........--------------------------....------.(((((((..((((........)))).(((((.(....)))))).....(((((.......))))))))))))... ( -30.80) >Bclau.0 4281557 115 + 4303871/0-120 GCCAUUUACAUCUUGAUAA-----AAAGUCGAAAGAAGCGAAAAUGGCUCGGUAGCUCAGUUGGUAGAGCAACGGACUGAAAAUCCGUGUGUCGGCGGUUCGAUUCCGUCCCGAGCCACC ((.........(((.....-----.)))((....)).)).....((((((((..((((........)))).(((((.(....)))))).....(((((.......))))))))))))).. ( -36.40) >Bsubt.0 4153807 114 + 4214630/0-120 GCCAUUUUACUUUACUUAG-----UAGAGAAAAAUAAA-ACAUUUGGCUCGGUAGCUCAGUUGGUAGAGCAACGGACUGAAAAUCCGUGUGUCGGCGGUUCGAUUCCGUCCCGAGCCACU ...(((((.((((((...)-----))))).)))))...-.....((((((((..((((........)))).(((((.(....)))))).....(((((.......))))))))))))).. ( -37.60) >Banth.0 5206672 120 + 5227293/0-120 GCCACUUUUGUUUAUACCACCAUUAGUGGUUUUAUAUAUAGUUUUGGCUCGGUAGCUCAGUCGGUAGAGCAGAGGACUGAAAAUCCUCGUGUCGGCGGUUCGAUUCCGUCCCGAGCCACU ((((((..((..........))..))))))..............((((((((..((((........)))).(((((.(....)))))).....(((((.......))))))))))))).. ( -41.50) >Bcere.0 5203671 120 + 5224283/0-120 GCCACUUUUGUUUAUACCACCAUUAGUGGUUUUAUAUAUAGUUUUGGCUCGGUAGCUCAGUCGGUAGAGCAGAGGACUGAAAAUCCUCGUGUCGGCGGUUCGAUUCCGUCCCGAGCCACU ((((((..((..........))..))))))..............((((((((..((((........)))).(((((.(....)))))).....(((((.......))))))))))))).. ( -41.50) >consensus GCCACUUU__UUUA_AUAA_____AAAGGU___AUAUA_AGAUUUGGCUCGGUAGCUCAGUCGGUAGAGCAACGGACUGAAAAUCCGUGUGUCGGCGGUUCGAUUCCGUCCCGAGCCACU .............................................(((((((..((((........)))).(((((.(....)))))).....(((((.......))))))))))))... (-33.34 = -32.28 + -1.05)

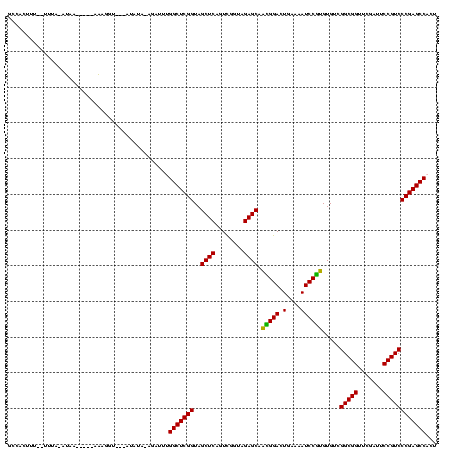
| Location | 4,187,073 – 4,187,178 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.84 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -29.99 |
| Energy contribution | -28.88 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
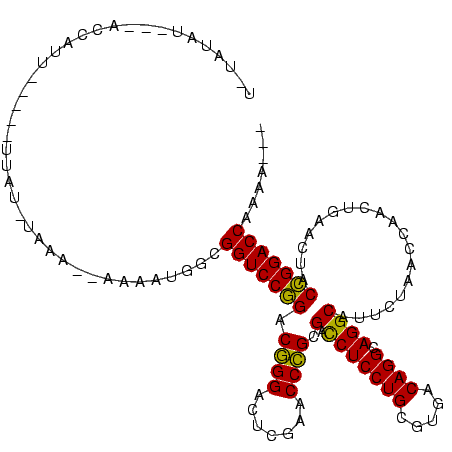
>Blich.0 4187073 105 + 4222334/80-200 UGUAUGU---AACUUU-----UUAU-------AAAAUGGCGGUCCGGACGGGACUCGAACCCGCGACCUCCUGCGUGACAGGCAGGCAUUCUAACCAACUGAACUACCGGACCUUGCAUG (((((((---((....-----))))-------).......(((((((.((((.......)))).(.(((((((.....)))).))))...................))))))).)))).. ( -30.30) >Bhalo.0 4163644 89 + 4202352/80-200 --UAUG--------------------------AAAGUGGCGGUCCGGACGGGACUCGAACCCGCGACCUCCUGCGUGACAGGCAGGCAUUCUAACCAACUGAACUACCGGACCUUAU--- --....--------------------------........(((((((.((((.......)))).(.(((((((.....)))).))))...................)))))))....--- ( -29.10) >Bclau.0 4281557 112 + 4303871/80-200 CGCUUCUUUCGACUUU-----UUAUCAAGAUGUAAAUGGCGGUCCGGACGGGACUCGAACCCGCGACCUCCUGCGUGACAGGCAGGCAUUCUAACCAACUGAACUACCGGACCAAGA--- .(((.....((.(((.-----.....))).)).....)))(((((((.((((.......)))).(.(((((((.....)))).))))...................)))))))....--- ( -33.30) >Bsubt.0 4153807 114 + 4214630/80-200 U-UUUAUUUUUCUCUA-----CUAAGUAAAGUAAAAUGGCGGUCCGGACGGGACUCGAACCCGCGACCUCCUGCGUGACAGGCAGGCAUUCUAACCAACUGAACUACCGGACCUUACAGU (-((((((((.((...-----...)).)))))))))....(((((((.((((.......)))).(.(((((((.....)))).))))...................)))))))....... ( -32.80) >Banth.0 5206672 116 + 5227293/80-200 UAUAUAUAAAACCACUAAUGGUGGUAUAAACAAAAGUGGCGGUCCCGACCGGGGUCGAACCGGCGAUCUCCUGCGUGACAGGCAGGCAUGUUAACCA-CUACACCACGGGACCAAAA--- ..........(((((.....)))))...............(((((((.((((.......))))......(((((.......)))))...........-........)))))))....--- ( -35.90) >Bcere.0 5203671 116 + 5224283/80-200 UAUAUAUAAAACCACUAAUGGUGGUAUAAACAAAAGUGGCGGUCCCGACCGGGGUCGAACCGGCGAUCUCCUGCGUGACAGGCAGGCAUGUUAACCA-CUACACCACGGGACCAAAA--- ..........(((((.....)))))...............(((((((.((((.......))))......(((((.......)))))...........-........)))))))....--- ( -35.90) >consensus U_UAUAU___ACCAUU_____UUAU_UAAA__AAAAUGGCGGUCCGGACGGGACUCGAACCCGCGACCUCCUGCGUGACAGGCAGGCAUUCUAACCAACUGAACUACCGGACCAAAA___ ........................................(((((((.((((.......)))).(.(((((((.....)))).))))...................)))))))....... (-29.99 = -28.88 + -1.11)
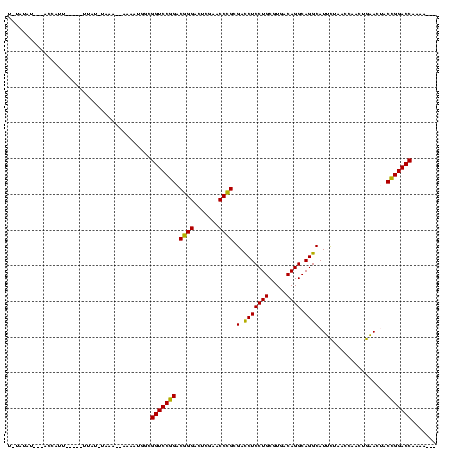
| Location | 4,187,073 – 4,187,178 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.84 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -31.47 |
| Energy contribution | -30.03 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 4187073 105 + 4222334/80-200 CAUGCAAGGUCCGGUAGUUCAGUUGGUUAGAAUGCCUGCCUGUCACGCAGGAGGUCGCGGGUUCGAGUCCCGUCCGGACCGCCAUUUU-------AUAA-----AAAGUU---ACAUACA .......(((((((...................((((.((((.....)))))))).(((((.......))))))))))))........-------....-----......---....... ( -32.30) >Bhalo.0 4163644 89 + 4202352/80-200 ---AUAAGGUCCGGUAGUUCAGUUGGUUAGAAUGCCUGCCUGUCACGCAGGAGGUCGCGGGUUCGAGUCCCGUCCGGACCGCCACUUU--------------------------CAUA-- ---....(((((((...................((((.((((.....)))))))).(((((.......))))))))))))........--------------------------....-- ( -32.30) >Bclau.0 4281557 112 + 4303871/80-200 ---UCUUGGUCCGGUAGUUCAGUUGGUUAGAAUGCCUGCCUGUCACGCAGGAGGUCGCGGGUUCGAGUCCCGUCCGGACCGCCAUUUACAUCUUGAUAA-----AAAGUCGAAAGAAGCG ---....(((((((...................((((.((((.....)))))))).(((((.......))))))))))))((.........(((.....-----.)))((....)).)). ( -36.10) >Bsubt.0 4153807 114 + 4214630/80-200 ACUGUAAGGUCCGGUAGUUCAGUUGGUUAGAAUGCCUGCCUGUCACGCAGGAGGUCGCGGGUUCGAGUCCCGUCCGGACCGCCAUUUUACUUUACUUAG-----UAGAGAAAAAUAAA-A .......(((((((...................((((.((((.....)))))))).(((((.......))))))))))))...(((((.((((((...)-----))))).)))))...-. ( -37.30) >Banth.0 5206672 116 + 5227293/80-200 ---UUUUGGUCCCGUGGUGUAG-UGGUUAACAUGCCUGCCUGUCACGCAGGAGAUCGCCGGUUCGACCCCGGUCGGGACCGCCACUUUUGUUUAUACCACCAUUAGUGGUUUUAUAUAUA ---....((((((.(((((..(-((.....))).(((((.......)))))....)))))....(((....)))))))))((((((..((..........))..)))))).......... ( -38.60) >Bcere.0 5203671 116 + 5224283/80-200 ---UUUUGGUCCCGUGGUGUAG-UGGUUAACAUGCCUGCCUGUCACGCAGGAGAUCGCCGGUUCGACCCCGGUCGGGACCGCCACUUUUGUUUAUACCACCAUUAGUGGUUUUAUAUAUA ---....((((((.(((((..(-((.....))).(((((.......)))))....)))))....(((....)))))))))((((((..((..........))..)))))).......... ( -38.60) >consensus ___UUAAGGUCCGGUAGUUCAGUUGGUUAGAAUGCCUGCCUGUCACGCAGGAGGUCGCGGGUUCGAGUCCCGUCCGGACCGCCACUUU__UUUA_AUAA_____AAAGGU___AUAUA_A .......(((((((..((((.........))))((((.((((.....)))))))).(((((.......))))))))))))........................................ (-31.47 = -30.03 + -1.44)

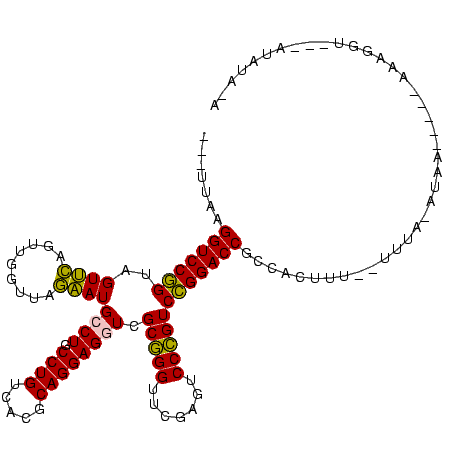
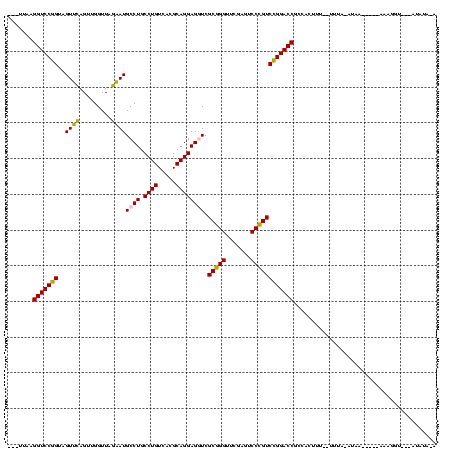
| Location | 4,187,073 – 4,187,193 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.83 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -37.65 |
| Energy contribution | -38.15 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 4187073 120 + 4222334/120-240 GGUCCGGACGGGACUCGAACCCGCGACCUCCUGCGUGACAGGCAGGCAUUCUAACCAACUGAACUACCGGACCUUGCAUGAUUACAGUACCUGCUGCGGAGAAGAAUCGUGGUCUCCUCC (((((((.((((.......)))).(.(((((((.....)))).))))...................)))))))...........((((....)))).((((.(((.......))).)))) ( -37.20) >Bsubt.0 4153807 113 + 4214630/120-240 GGUCCGGACGGGACUCGAACCCGCGACCUCCUGCGUGACAGGCAGGCAUUCUAACCAACUGAACUACCGGACCUUACAGUA-------AUUGCCCGGAGAGCUGUCGUGUCAGCUCUCAC (((((((.((((.......)))).(.(((((((.....)))).))))...................)))))))........-------........((((((((......)))))))).. ( -43.20) >consensus GGUCCGGACGGGACUCGAACCCGCGACCUCCUGCGUGACAGGCAGGCAUUCUAACCAACUGAACUACCGGACCUUACAGGA_______ACCGCCCGCAGAGAAGAAGCGUCAGCUCCCAC (((((((.((((.......)))).(.(((((((.....)))).))))...................))))))).......................((((((((......)))))))).. (-37.65 = -38.15 + 0.50)

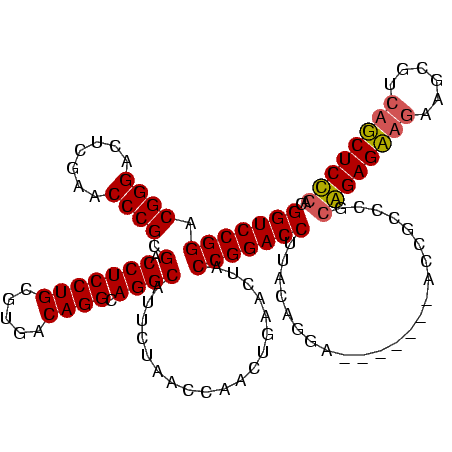
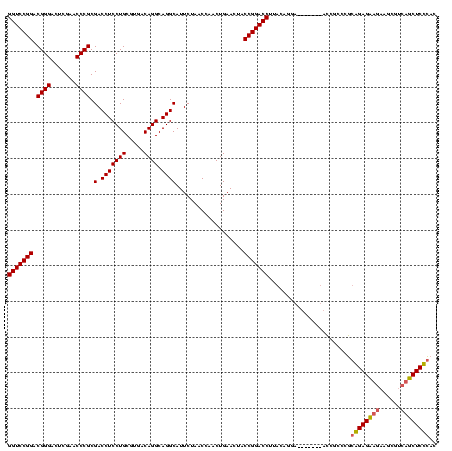
| Location | 4,187,073 – 4,187,193 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.00 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -23.00 |
| Energy contribution | -25.25 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 4187073 120 + 4222334/160-280 UGAUUUACUUUAGCGUUAUUGCUAAAUCCCCGCUUGCUCCGGAGGAGACCACGAUUCUUCUCCGCAGCAGGUACUGUAAUCAUGCAAGGUCCGGUAGUUCAGUUGGUUAGAAUGCCUGCC (((.(((((((((((....))))))....(((((((((.(((((((((.......))))))))).)))))))..((((....)))).))...))))).)))((.(((......))).)). ( -39.50) >Bsubt.0 4153807 113 + 4214630/160-280 UGAUUUACUUUAGCGUUAUUGCUAAAUUCCUUAUUUGUCUGUGAGAGCUGACACGACAGCUCUCCGGGCAAU-------UACUGUAAGGUCCGGUAGUUCAGUUGGUUAGAAUGCCUGCC (((.(((((((((((....))))))...(((((((((((((.((((((((......))))))))))))))).-------....))))))...))))).)))((.(((......))).)). ( -40.30) >consensus UGAUUUACUUUAGCGUUAUUGCUAAAUCCCCGAUUGCUCCGGAAGAGACCACAAGACAGCUCCCCAGCAAAU_______UAAUGCAAGGUCCGGUAGUUCAGUUGGUUAGAAUGCCUGCC (((.(((((((((((....)))))).........(((((((.((((((((......))))))))))))))).....................))))).)))((.(((......))).)). (-23.00 = -25.25 + 2.25)

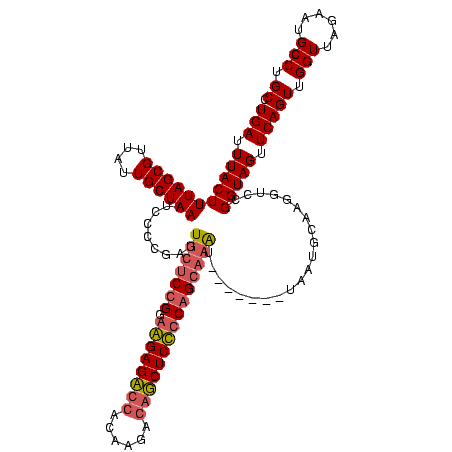
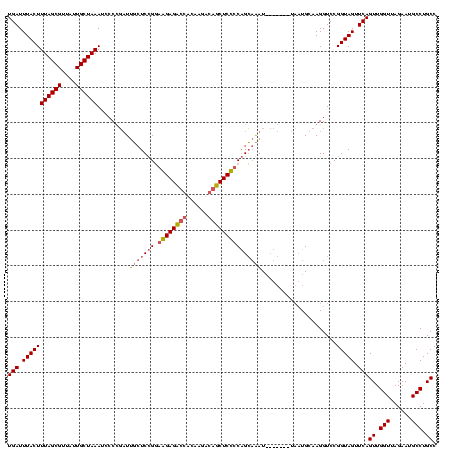
| Location | 4,187,073 – 4,187,193 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.50 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -24.15 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 4187073 120 + 4222334/200-320 AUUACAGUACCUGCUGCGGAGAAGAAUCGUGGUCUCCUCCGGAGCAAGCGGGGAUUUAGCAAUAACGCUAAAGUAAAUCAAUGACCCGUACGGGAUUCGAACCCGUGUUACCGCCGUGAA .((((......((((.(((((.(((.......))).))))).)))).((((...((((((......))))))................((((((.......))))))...)))).)))). ( -39.80) >Bsubt.0 4153807 113 + 4214630/200-320 A-------AUUGCCCGGAGAGCUGUCGUGUCAGCUCUCACAGACAAAUAAGGAAUUUAGCAAUAACGCUAAAGUAAAUCAAUGACCCGUACGGGAUUCGAACCCGUGUUACCGCCGUGAA .-------.(..(...((((((((......))))))))............((..((((((......))))))................((((((.......))))))...))...)..). ( -30.60) >consensus A_______ACCGCCCGCAGAGAAGAAGCGUCAGCUCCCACAGACAAAGAAGGAAUUUAGCAAUAACGCUAAAGUAAAUCAAUGACCCGUACGGGAUUCGAACCCGUGUUACCGCCGUGAA .........(((....((((((((......)))))))).)))........((..((((((......))))))................((((((.......))))))...))........ (-24.15 = -24.40 + 0.25)

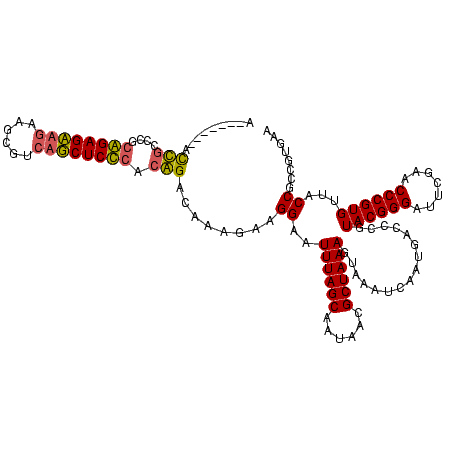
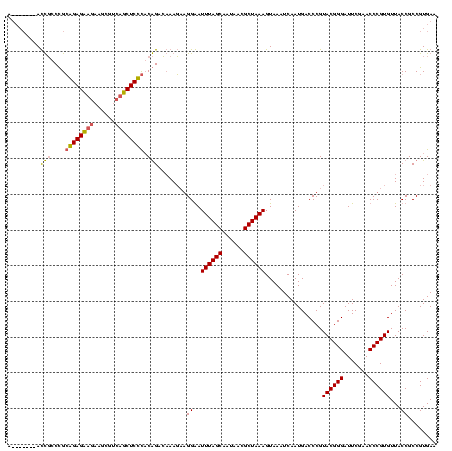
| Location | 4,187,073 – 4,187,193 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.50 |
| Mean single sequence MFE | -43.20 |
| Consensus MFE | -27.70 |
| Energy contribution | -29.95 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.64 |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 4187073 120 + 4222334/200-320 UUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGGUCAUUGAUUUACUUUAGCGUUAUUGCUAAAUCCCCGCUUGCUCCGGAGGAGACCACGAUUCUUCUCCGCAGCAGGUACUGUAAU ......((((((...((((.((((..(((....)))...)))).....(((((((....)))))))..))))((((((.(((((((((.......))))))))).))))))))))))... ( -45.80) >Bsubt.0 4153807 113 + 4214630/200-320 UUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGGUCAUUGAUUUACUUUAGCGUUAUUGCUAAAUUCCUUAUUUGUCUGUGAGAGCUGACACGACAGCUCUCCGGGCAAU-------U .....((((((((((((((.......)))))..(((((...)))))........)))))))))...........(((((((.((((((((......))))))))))))))).-------. ( -40.60) >consensus UUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGGUCAUUGAUUUACUUUAGCGUUAUUGCUAAAUCCCCGAUUGCUCCGGAAGAGACCACAAGACAGCUCCCCAGCAAAU_______U .....((((((((((((((.......)))))..(((((...)))))........)))))))))...........(((((((.((((((((......)))))))))))))))......... (-27.70 = -29.95 + 2.25)

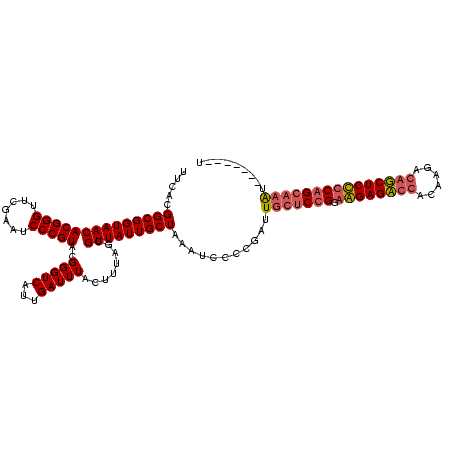
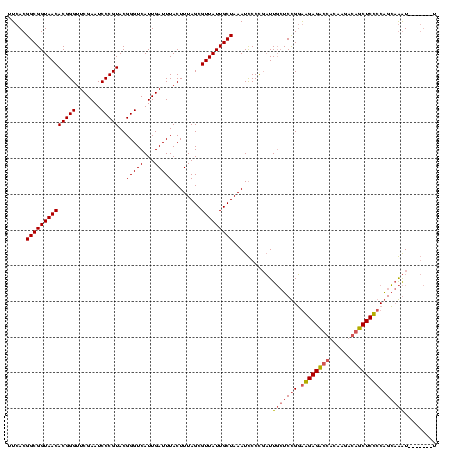
| Location | 4,187,073 – 4,187,193 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.45 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -26.76 |
| Energy contribution | -26.96 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 4187073 120 + 4222334/236-356 CUCCGGAGCAAGCGGGGAUUUAGCAAUAACGCUAAAGUAAAUCAAUGACCCGUACGGGAUUCGAACCCGUGUUACCGCCGUGAAAGGGCGGUGUCUUAACCACUUGACCAACGGGCCAGG (((((.......))))).((((((......)))))).........((.((((((((((.......)))))..(((((((.......))))))).................))))).)).. ( -39.00) >Bclau.0 4281557 94 + 4303871/236-356 ------------UAUGUAUCUAUAUGAAA--------------AGUGACCCGUACGGGAUUCGAACCCGUGUUACCGCCGUGAAAGGGCGGUGUCUUAACCGCUUGACCAACGGGCCACU ------------.................--------------((((.((((((((((.......)))))..(((((((.......))))))).................))))).)))) ( -31.30) >Bsubt.0 4153807 120 + 4214630/236-356 UCACAGACAAAUAAGGAAUUUAGCAAUAACGCUAAAGUAAAUCAAUGACCCGUACGGGAUUCGAACCCGUGUUACCGCCGUGAAAGGGCGGUGUCUUAACCGCUUGACCAACGGGCCUUC ............((((..((((((......))))))............((((((((((.......)))))..(((((((.......))))))).................))))))))). ( -34.50) >Banth.0 5206672 91 + 5227293/236-356 -------------AUGAACAUAAAAAA----------------CAUGACCCGUACGGGAUUCGAACCCGUGUUACCGCCGUGAAAGGGCGGUGUCUUAACCACUUGACCAACGGGCCACG -------------..............----------------..((.((((((((((.......)))))..(((((((.......))))))).................))))).)).. ( -27.50) >Bcere.0 5203671 92 + 5224283/236-356 -------------AUGAACAUAAAAAAA---------------CAUGACCCGUACGGGAUUCGAACCCGUGUUACCGCCGUGAAAGGGCGGUGUCUUAACCACUUGACCAACGGGCCACG -------------...............---------------..((.((((((((((.......)))))..(((((((.......))))))).................))))).)).. ( -27.50) >consensus _____________AUGAAUAUAAAAAAAA______________AAUGACCCGUACGGGAUUCGAACCCGUGUUACCGCCGUGAAAGGGCGGUGUCUUAACCACUUGACCAACGGGCCACG .............................................((.((((((((((.......)))))..(((((((.......))))))).................))))).)).. (-26.76 = -26.96 + 0.20)

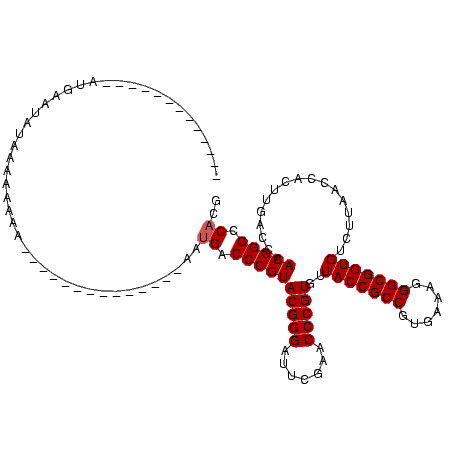
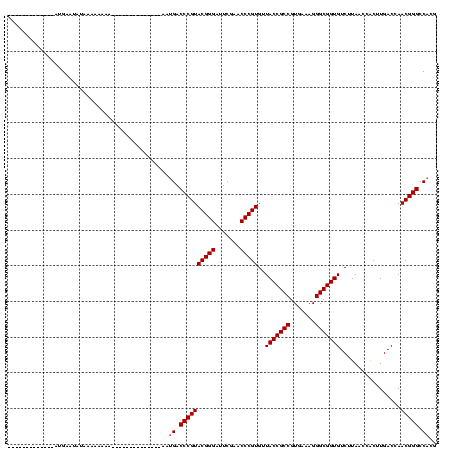
| Location | 4,187,073 – 4,187,193 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.45 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -32.46 |
| Energy contribution | -32.46 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 4187073 120 + 4222334/236-356 CCUGGCCCGUUGGUCAAGUGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGGUCAUUGAUUUACUUUAGCGUUAUUGCUAAAUCCCCGCUUGCUCCGGAG ......(((..((.(((((((....(((((((((.......))))))...(((((.......)))))....)))..........(((((((....)))))))...))))))))).))).. ( -41.10) >Bclau.0 4281557 94 + 4303871/236-356 AGUGGCCCGUUGGUCAAGCGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGGUCACU--------------UUUCAUAUAGAUACAUA------------ ((((((((((..(((..........)))((((((.......))))))...(((((.......)))))))))))))))--------------.................------------ ( -34.70) >Bsubt.0 4153807 120 + 4214630/236-356 GAAGGCCCGUUGGUCAAGCGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGGUCAUUGAUUUACUUUAGCGUUAUUGCUAAAUUCCUUAUUUGUCUGUGA ((((((((((..(((..........)))((((((.......))))))...(((((.......))))))))))))..........(((((((....))))))))))............... ( -36.40) >Banth.0 5206672 91 + 5227293/236-356 CGUGGCCCGUUGGUCAAGUGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGGUCAUG----------------UUUUUUAUGUUCAU------------- ((((((((((..(((..........)))((((((.......))))))...(((((.......)))))))))))))))----------------..............------------- ( -34.20) >Bcere.0 5203671 92 + 5224283/236-356 CGUGGCCCGUUGGUCAAGUGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGGUCAUG---------------UUUUUUUAUGUUCAU------------- ((((((((((..(((..........)))((((((.......))))))...(((((.......)))))))))))))))---------------...............------------- ( -34.20) >consensus CGUGGCCCGUUGGUCAAGUGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGGUCAUU______________UUUUUUCUAAAUUCAU_____________ ((((((((((..(((..........)))((((((.......))))))...(((((.......)))))))))))))))........................................... (-32.46 = -32.46 + 0.00)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:16:51 2006