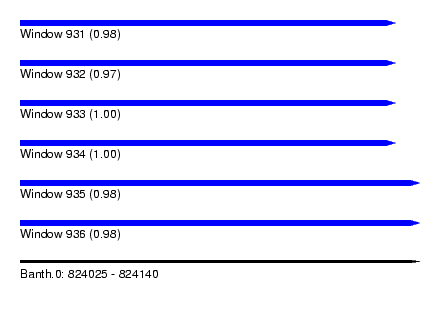
| Sequence ID | Banth.0 |
|---|---|
| Location | 824,025 – 824,140 |
| Length | 115 |
| Max. P | 0.999924 |

| Location | 824,025 – 824,133 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.76 |
| Mean single sequence MFE | -43.78 |
| Consensus MFE | -35.96 |
| Energy contribution | -35.96 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.97 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 824025 108 + 5227293/0-120 AAGGGGAGUAACUUAUUACAGUAAAGUCGUCAUUACAGGGA--------GAAACCCUCGGCUUUAUUGGCAACG----UUUCGUUGUUAGUGAGACCUUUACCAGCAAUGGUAAAGGCCC ..(((......((((((((((((((((((.......((((.--------....)))))))))))))))((((((----...))))))))))))).(((((((((....)))))))))))) ( -43.61) >Bcere.0 926777 108 + 5224283/0-120 AAGGGGAGUAACUUAUUACAGUAAAGUCGUCAUUACAGGGA--------GAAACCCUCGGCUUUAUUGGCAACG----UUUCGUUGUUAGUGAGACCUUUACCAGCAAUGGUAAAGGCCC ..(((......((((((((((((((((((.......((((.--------....)))))))))))))))((((((----...))))))))))))).(((((((((....)))))))))))) ( -43.61) >Bhalo.0 1524417 120 - 4202352/0-120 AAGGGGAGUAGCUAAUUACAAUAAAGUCGUCAUGACAGGGAUCUCCCAUAAAUCCCUCGGCUUUAUUGGCAACGGAUAUACGGUUGUUAGCAAGACCUUUACCAAAUGGGGUAAAGGUCU ..........((((....(((((((((((.......((((((.........)))))))))))))))))(((((.(.....).))))))))).(((((((((((......))))))))))) ( -44.11) >consensus AAGGGGAGUAACUUAUUACAGUAAAGUCGUCAUUACAGGGA________GAAACCCUCGGCUUUAUUGGCAACG____UUUCGUUGUUAGUGAGACCUUUACCAGCAAUGGUAAAGGCCC ..(((......((((((((((((((((((.......((((.............)))))))))))))))(((((.........)))))))))))).((((((((......))))))))))) (-35.96 = -35.96 + 0.01)

| Location | 824,025 – 824,133 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.76 |
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -26.71 |
| Energy contribution | -26.16 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 824025 108 + 5227293/0-120 GGGCCUUUACCAUUGCUGGUAAAGGUCUCACUAACAACGAAA----CGUUGCCAAUAAAGCCGAGGGUUUC--------UCCCUGUAAUGACGACUUUACUGUAAUAAGUUACUCCCCUU ((((((((((((....))))))))))))......(((((...----)))))............((((....--------.))))(((((....((......)).....)))))....... ( -28.20) >Bcere.0 926777 108 + 5224283/0-120 GGGCCUUUACCAUUGCUGGUAAAGGUCUCACUAACAACGAAA----CGUUGCCAAUAAAGCCGAGGGUUUC--------UCCCUGUAAUGACGACUUUACUGUAAUAAGUUACUCCCCUU ((((((((((((....))))))))))))......(((((...----)))))............((((....--------.))))(((((....((......)).....)))))....... ( -28.20) >Bhalo.0 1524417 120 - 4202352/0-120 AGACCUUUACCCCAUUUGGUAAAGGUCUUGCUAACAACCGUAUAUCCGUUGCCAAUAAAGCCGAGGGAUUUAUGGGAGAUCCCUGUCAUGACGACUUUAUUGUAAUUAGCUACUCCCCUU (((((((((((......))))))))))).(((((((((.(.....).)))).((((((((.(.((((((((.....))))))))((....))).))))))))...))))).......... ( -38.30) >consensus GGGCCUUUACCAUUGCUGGUAAAGGUCUCACUAACAACGAAA____CGUUGCCAAUAAAGCCGAGGGUUUC________UCCCUGUAAUGACGACUUUACUGUAAUAAGUUACUCCCCUU (((((((((((......))))))))))).(((..(((((.......))))).((.(((((.((((((.............)))).......)).))))).)).....))).......... (-26.71 = -26.16 + -0.55)

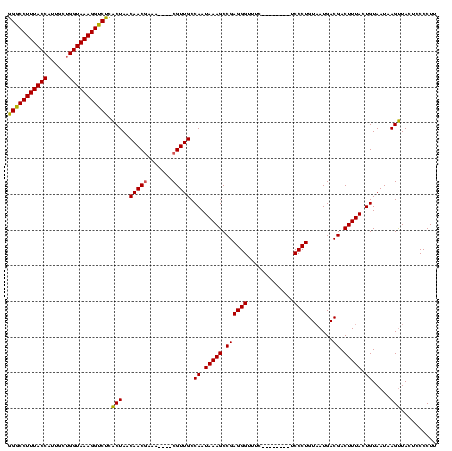
| Location | 824,025 – 824,133 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -40.24 |
| Consensus MFE | -31.71 |
| Energy contribution | -30.83 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.17 |
| Mean z-score | -4.55 |
| Structure conservation index | 0.79 |
| SVM decision value | 4.58 |
| SVM RNA-class probability | 0.999924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 824025 108 + 5227293/40-160 A--------GAAACCCUCGGCUUUAUUGGCAACG----UUUCGUUGUUAGUGAGACCUUUACCAGCAAUGGUAAAGGCCCCUUAUUGUCUUUUUUAAGACCCUUACCAUAUUUGGUAGGG .--------.........((((((((((((((((----...)))))))))))))..((((((((....))))))))))).......(((((....)))))((((((((....)))))))) ( -41.30) >Bcere.0 926777 108 + 5224283/40-160 A--------GAAACCCUCGGCUUUAUUGGCAACG----UUUCGUUGUUAGUGAGACCUUUACCAGCAAUGGUAAAGGCCCCUUAUUGUCUUUUUUAGGACCCUUACCAUAUUUGGUAGGG .--------.........((((((((((((((((----...)))))))))))))..((((((((....))))))))))).......(((((....)))))((((((((....)))))))) ( -41.00) >Bhalo.0 1524417 119 - 4202352/40-160 AUCUCCCAUAAAUCCCUCGGCUUUAUUGGCAACGGAUAUACGGUUGUUAGCAAGACCUUUACCAAAUGGGGUAAAGGUCUUUUCCUAUGCUUUUUUAGACCUUUGCC-UAUGGGGCAAAG ...(((((((.((((....(((.....)))...))))....((((...(((((((((((((((......))))))))))).......))))......))))......-)))))))..... ( -38.41) >consensus A________GAAACCCUCGGCUUUAUUGGCAACG____UUUCGUUGUUAGUGAGACCUUUACCAGCAAUGGUAAAGGCCCCUUAUUGUCUUUUUUAAGACCCUUACCAUAUUUGGUAGGG ..................(((((((((((((((.........))))))))))))..(((((((......)))))))))).....................((((((((....)))))))) (-31.71 = -30.83 + -0.88)

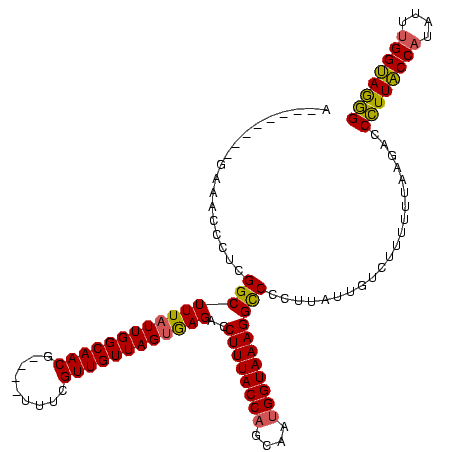
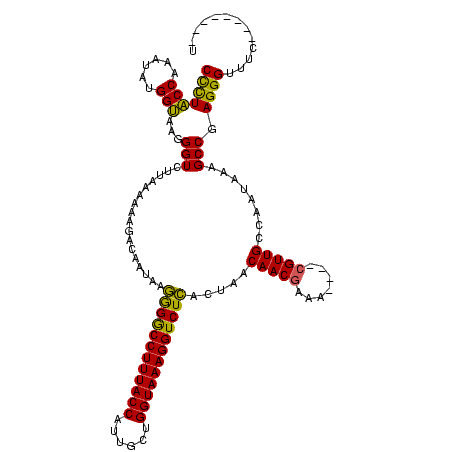
| Location | 824,025 – 824,133 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -39.39 |
| Consensus MFE | -32.13 |
| Energy contribution | -30.70 |
| Covariance contribution | -1.43 |
| Combinations/Pair | 1.22 |
| Mean z-score | -4.83 |
| Structure conservation index | 0.82 |
| SVM decision value | 4.09 |
| SVM RNA-class probability | 0.999791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 824025 108 + 5227293/40-160 CCCUACCAAAUAUGGUAAGGGUCUUAAAAAAGACAAUAAGGGGCCUUUACCAUUGCUGGUAAAGGUCUCACUAACAACGAAA----CGUUGCCAAUAAAGCCGAGGGUUUC--------U ((((........((((((.((((((....))))).....(((((((((((((....))))))))))))).............----).)))))).........))))....--------. ( -37.93) >Bcere.0 926777 108 + 5224283/40-160 CCCUACCAAAUAUGGUAAGGGUCCUAAAAAAGACAAUAAGGGGCCUUUACCAUUGCUGGUAAAGGUCUCACUAACAACGAAA----CGUUGCCAAUAAAGCCGAGGGUUUC--------U ((((((((...((((((((((((((..............))))))))))))))...))))...(((........(((((...----)))))........))).))))....--------. ( -36.43) >Bhalo.0 1524417 119 - 4202352/40-160 CUUUGCCCCAUA-GGCAAAGGUCUAAAAAAGCAUAGGAAAAGACCUUUACCCCAUUUGGUAAAGGUCUUGCUAACAACCGUAUAUCCGUUGCCAAUAAAGCCGAGGGAUUUAUGGGAGAU (((((((.....-)))))))((((.........(((...((((((((((((......)))))))))))).)))....(((((.((((.(((.(......).))).)))).))))).)))) ( -43.80) >consensus CCCUACCAAAUAUGGUAAGGGUCUUAAAAAAGACAAUAAGGGGCCUUUACCAUUGCUGGUAAAGGUCUCACUAACAACGAAA____CGUUGCCAAUAAAGCCGAGGGUUUC________U (((((((......)))...(((.................((((((((((((......)))))))))))).....(((((.......)))))........))).))))............. (-32.13 = -30.70 + -1.43)

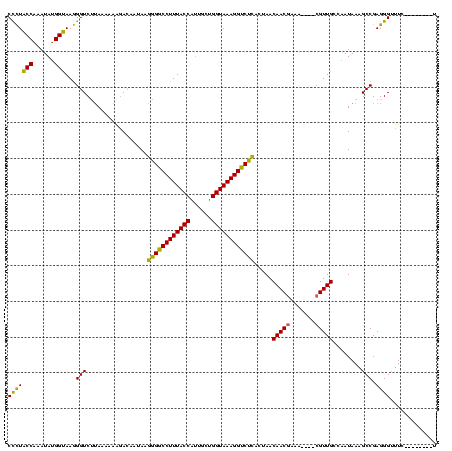
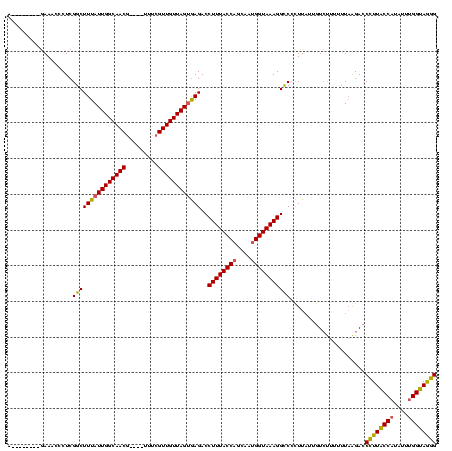
| Location | 824,025 – 824,140 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -45.67 |
| Consensus MFE | -36.50 |
| Energy contribution | -35.40 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.18 |
| Mean z-score | -5.93 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 824025 115 + 5227293/48-168 -GAAACCCUCGGCUUUAUUGGCAACG----UUUCGUUGUUAGUGAGACCUUUACCAGCAAUGGUAAAGGCCCCUUAUUGUCUUUUUUAAGACCCUUACCAUAUUUGGUAGGGGCCUUUUU -.........((..((((((((((((----...))))))))))))..))(((((((....)))))))((((((.....(((((....)))))...(((((....)))))))))))..... ( -46.00) >Bcere.0 926777 115 + 5224283/48-168 -GAAACCCUCGGCUUUAUUGGCAACG----UUUCGUUGUUAGUGAGACCUUUACCAGCAAUGGUAAAGGCCCCUUAUUGUCUUUUUUAGGACCCUUACCAUAUUUGGUAGGGGCCUUUUU -.........((((((((((((((((----...))))))))))))).(((((((((....))))))))).........)))......(((.(((((((((....)))))))))))).... ( -45.80) >Bhalo.0 1524417 119 - 4202352/48-168 UAAAUCCCUCGGCUUUAUUGGCAACGGAUAUACGGUUGUUAGCAAGACCUUUACCAAAUGGGGUAAAGGUCUUUUCCUAUGCUUUUUUAGACCUUUGCC-UAUGGGGCAAAGGUCUUUUU ..........(((....((((((((.(.....).)))))))).((((((((((((......)))))))))))).......))).....(((((((((((-.....))))))))))).... ( -45.20) >consensus _GAAACCCUCGGCUUUAUUGGCAACG____UUUCGUUGUUAGUGAGACCUUUACCAGCAAUGGUAAAGGCCCCUUAUUGUCUUUUUUAAGACCCUUACCAUAUUUGGUAGGGGCCUUUUU ..........(((((((((((((((.........))))))))))))..(((((((......)))))))))).................((.(((((((((....))))))))).)).... (-36.50 = -35.40 + -1.10)

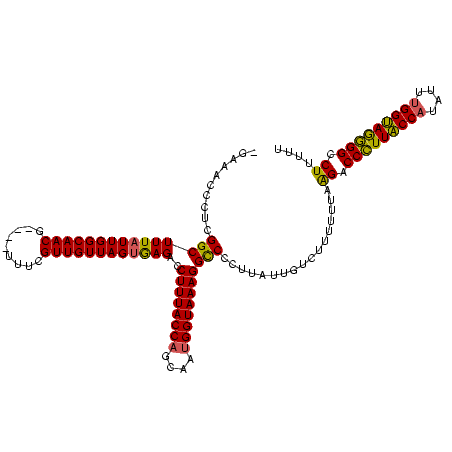
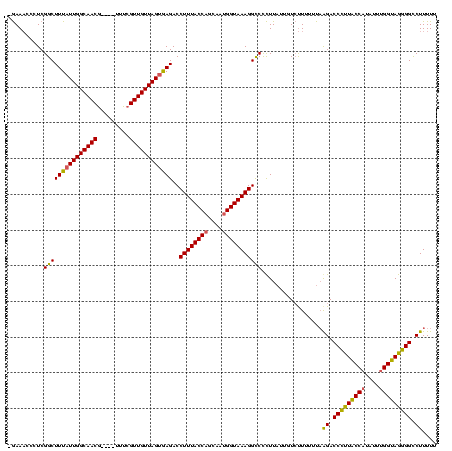
| Location | 824,025 – 824,140 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -41.23 |
| Consensus MFE | -37.09 |
| Energy contribution | -35.77 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -5.59 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 824025 115 + 5227293/48-168 AAAAAGGCCCCUACCAAAUAUGGUAAGGGUCUUAAAAAAGACAAUAAGGGGCCUUUACCAUUGCUGGUAAAGGUCUCACUAACAACGAAA----CGUUGCCAAUAAAGCCGAGGGUUUC- ...(((((((.(((((....))))).)))))))..............(((((((((((((....))))))))))))).....(((((...----)))))......(((((...))))).- ( -41.20) >Bcere.0 926777 115 + 5224283/48-168 AAAAAGGCCCCUACCAAAUAUGGUAAGGGUCCUAAAAAAGACAAUAAGGGGCCUUUACCAUUGCUGGUAAAGGUCUCACUAACAACGAAA----CGUUGCCAAUAAAGCCGAGGGUUUC- ....((((((.(((((....))))).))).)))..............(((((((((((((....))))))))))))).....(((((...----)))))......(((((...))))).- ( -39.20) >Bhalo.0 1524417 119 - 4202352/48-168 AAAAAGACCUUUGCCCCAUA-GGCAAAGGUCUAAAAAAGCAUAGGAAAAGACCUUUACCCCAUUUGGUAAAGGUCUUGCUAACAACCGUAUAUCCGUUGCCAAUAAAGCCGAGGGAUUUA ....(((((((((((.....-))))))))))).........(((...((((((((((((......)))))))))))).)))..........((((.(((.(......).))).))))... ( -43.30) >consensus AAAAAGGCCCCUACCAAAUAUGGUAAGGGUCUUAAAAAAGACAAUAAGGGGCCUUUACCAUUGCUGGUAAAGGUCUCACUAACAACGAAA____CGUUGCCAAUAAAGCCGAGGGUUUC_ .....(((((.((((......)))).)))))................((((((((((((......)))))))))))).....(((((.......)))))......(((((...))))).. (-37.09 = -35.77 + -1.32)

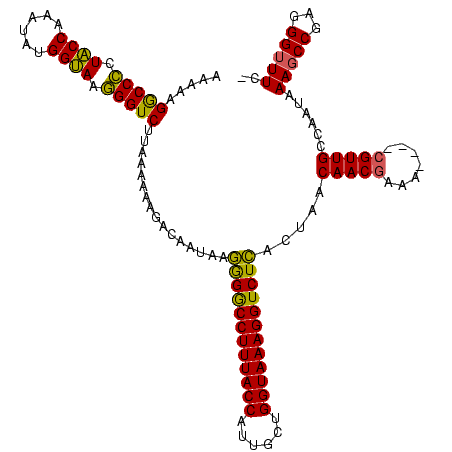
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:27:46 2006