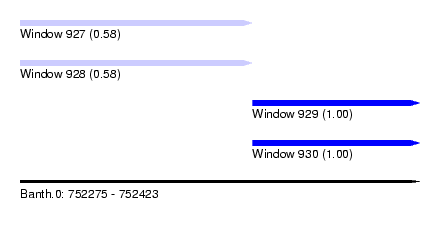
| Sequence ID | Banth.0 |
|---|---|
| Location | 752,275 – 752,423 |
| Length | 148 |
| Max. P | 0.998276 |

| Location | 752,275 – 752,361 |
|---|---|
| Length | 86 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -11.04 |
| Energy contribution | -11.04 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
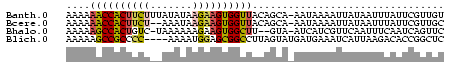
>Banth.0 752275 86 + 5227293 CACUUCCCUUCGCUAGUAUUACCUAGAUCAGGUGUUAUAAGGGUUAAG--GAU--UAUUCCUCUCUCAGC--ACAACAAGCACCCCUAGUGG ..........((((((...(((((.....)))))......((((..((--((.--...))))......((--.......)))))))))))). ( -19.90) >Bcere.0 828954 85 + 5224283 CACUUCCCUUCGCUAGUAUUACCUAGAUCAGGU-CUGUAAGGGUUAAG--GAU--UAUUCCUCUCUCAGC--ACAACAAGCACCCCUAGUGG ..........(((((((((.((((.....))))-..))).((((..((--((.--...))))......((--.......)))))))))))). ( -19.00) >Bclau.0 2048189 82 + 4303871 CACUUCCCUUCGCUAGCAUUACCUAGAUCAGGU----AAAGGGUUAAG--GCU-AAACGCCUUUCUCAGC--AUAA-ACGCACCCCUAGUGG ..........((((((..((((((.....))))----)).((((.(((--((.-....))))).....((--....-..)))))))))))). ( -23.80) >Bhalo.0 2798928 89 + 4202352 CACUUCCCUUCGCUAGUAUUACCUAGAUCAGGU-GAAUGAGGGUUAAGAUGCUAAAAAACAUCUCUCUCA--GCGGUUUGCACCCCUAGUGG ..........((((((..((((((.....))))-)).((((((...(((((........)))))))))))--..(((....))).)))))). ( -23.30) >Blich.0 185419 87 - 4222334 CACUUCCCUACGCUAGUAUGAACUAGAUCAGGU--UCAAAGGGUCCAA--GAU-CGCUUCUUGUCUCAGCCGGCUUACGGCUCCCCUAGUGG ..........((((((..((((((......)))--)))..(((..(((--((.-....)))))....(((((.....)))))))))))))). ( -28.40) >consensus CACUUCCCUUCGCUAGUAUUACCUAGAUCAGGU__UAUAAGGGUUAAG__GAU__AAUUCCUCUCUCAGC__ACAACAAGCACCCCUAGUGG ((((..(((...((((......))))...))).......((((........................................)))))))). (-11.04 = -11.04 + 0.00)

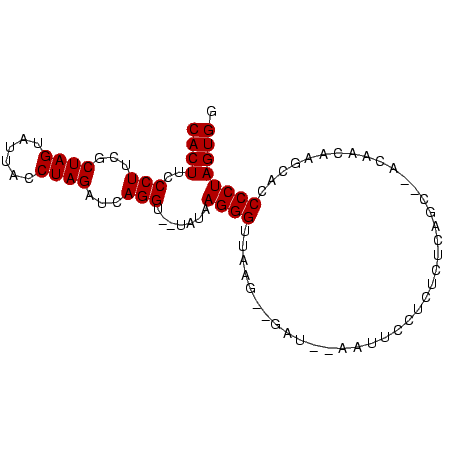
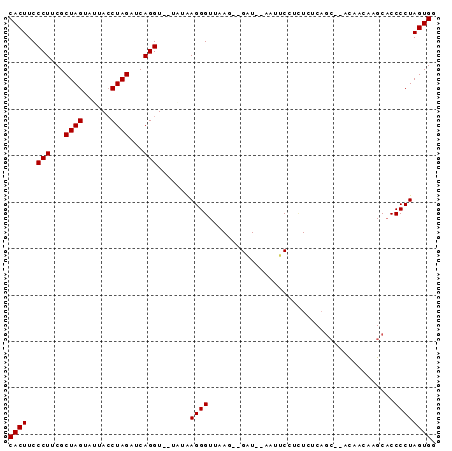
| Location | 752,275 – 752,361 |
|---|---|
| Length | 86 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -11.04 |
| Energy contribution | -11.04 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 752275 86 + 5227293/0-92 CACUUCCCUUCGCUAGUAUUACCUAGAUCAGGUGUUAUAAGGGUUAAG--GAU--UAUUCCUCUCUCAGC--ACAACAAGCACCCCUAGUGG ..........((((((...(((((.....)))))......((((..((--((.--...))))......((--.......)))))))))))). ( -19.90) >Bcere.0 828954 85 + 5224283/0-92 CACUUCCCUUCGCUAGUAUUACCUAGAUCAGGU-CUGUAAGGGUUAAG--GAU--UAUUCCUCUCUCAGC--ACAACAAGCACCCCUAGUGG ..........(((((((((.((((.....))))-..))).((((..((--((.--...))))......((--.......)))))))))))). ( -19.00) >Bclau.0 2048189 82 + 4303871/0-92 CACUUCCCUUCGCUAGCAUUACCUAGAUCAGGU----AAAGGGUUAAG--GCU-AAACGCCUUUCUCAGC--AUAA-ACGCACCCCUAGUGG ..........((((((..((((((.....))))----)).((((.(((--((.-....))))).....((--....-..)))))))))))). ( -23.80) >Bhalo.0 2798928 89 + 4202352/0-92 CACUUCCCUUCGCUAGUAUUACCUAGAUCAGGU-GAAUGAGGGUUAAGAUGCUAAAAAACAUCUCUCUCA--GCGGUUUGCACCCCUAGUGG ..........((((((..((((((.....))))-)).((((((...(((((........)))))))))))--..(((....))).)))))). ( -23.30) >Blich.0 185419 87 - 4222334/0-92 CACUUCCCUACGCUAGUAUGAACUAGAUCAGGU--UCAAAGGGUCCAA--GAU-CGCUUCUUGUCUCAGCCGGCUUACGGCUCCCCUAGUGG ..........((((((..((((((......)))--)))..(((..(((--((.-....)))))....(((((.....)))))))))))))). ( -28.40) >consensus CACUUCCCUUCGCUAGUAUUACCUAGAUCAGGU__UAUAAGGGUUAAG__GAU__AAUUCCUCUCUCAGC__ACAACAAGCACCCCUAGUGG ((((..(((...((((......))))...))).......((((........................................)))))))). (-11.04 = -11.04 + 0.00)

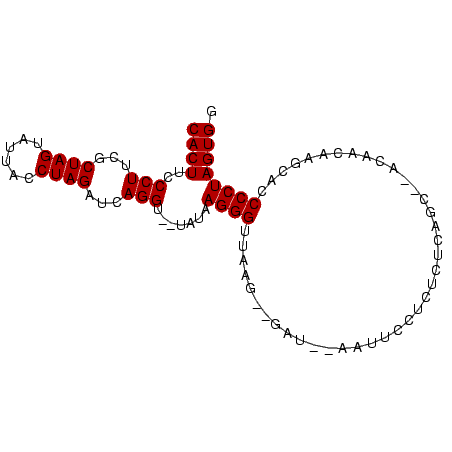
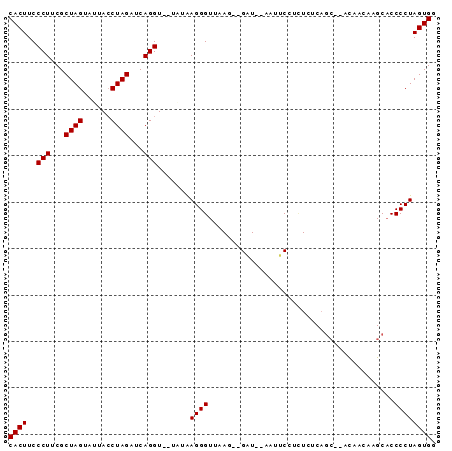
| Location | 752,361 – 752,423 |
|---|---|
| Length | 62 |
| Sequences | 4 |
| Columns | 63 |
| Reading direction | forward |
| Mean pairwise identity | 59.84 |
| Mean single sequence MFE | -18.67 |
| Consensus MFE | -13.18 |
| Energy contribution | -12.30 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.60 |
| Mean z-score | -4.36 |
| Structure conservation index | 0.71 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998083 |
| Prediction | RNA |
| WARNING | Sequence 1: Base composition out of range. |
Download alignment: ClustalW | MAF
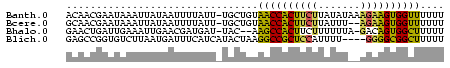
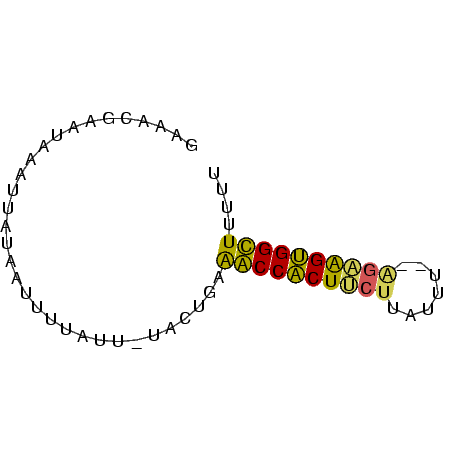
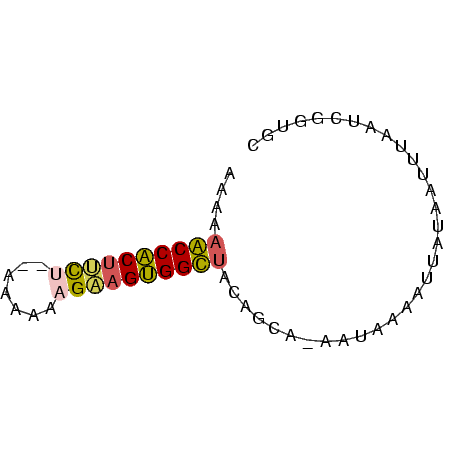
>Banth.0 752361 62 + 5227293 ACAACGAAUAAAUUAUAAUUUUAUU-UGCUGUAACCACUUCUUAUAUAAAGAAGUGGUUUUUU (((.((((((((.......))))))-)).)))(((((((((((.....))))))))))).... ( -20.20) >Bcere.0 829039 60 + 5224283 GCAACGAAUAAAUUAUAAUUUUAUU-UGCUGUAACCACUUCUUAUUU--AGAAGUGGUUUUUU (((.((((((((.......))))))-)).)))((((((((((.....--)))))))))).... ( -19.90) >Bhalo.0 2799043 59 + 4202352 GAACUGAUUGAAAUUGAACGAUGAU-UAC--AAGCCACUUCUUUUUUA-GACAGUGGCUUUUU .........................-...--(((((((((((.....)-)).))))))))... ( -12.80) >Blich.0 185506 59 - 4222334 GAGCCGGUGUCUUAAUGAUUUCAUCAUACUAAGGCCGCUCCAUUUU----GGGGCGGCUUUUU (((((.(.(((((((((((...)))))..)))))))(((((.....----))))))))))... ( -21.80) >consensus GAAACGAAUAAAUUAUAAUUUUAUU_UACUGAAACCACUUCUUAUUU__AGAAGUGGCUUUUU ................................((((((((((.......)))))))))).... (-13.18 = -12.30 + -0.88)
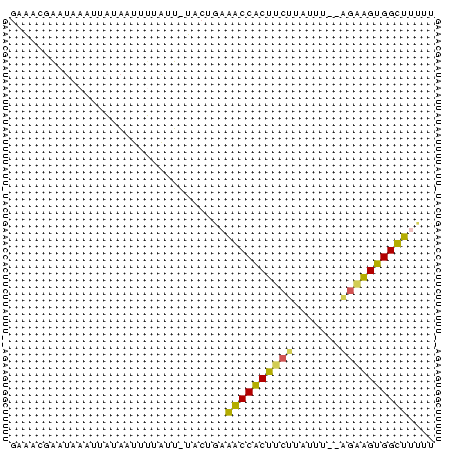
| Location | 752,361 – 752,423 |
|---|---|
| Length | 62 |
| Sequences | 4 |
| Columns | 63 |
| Reading direction | reverse |
| Mean pairwise identity | 59.84 |
| Mean single sequence MFE | -16.80 |
| Consensus MFE | -11.19 |
| Energy contribution | -11.12 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.40 |
| Mean z-score | -4.50 |
| Structure conservation index | 0.67 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998276 |
| Prediction | RNA |
| WARNING | Sequence 1: Base composition out of range. |
Download alignment: ClustalW | MAF
>Banth.0 752361 62 + 5227293 AAAAAACCACUUCUUUAUAUAAGAAGUGGUUACAGCA-AAUAAAAUUAUAAUUUAUUCGUUGU ....(((((((((((.....)))))))))))(((((.-((((((.......)))))).))))) ( -20.30) >Bcere.0 829039 60 + 5224283 AAAAAACCACUUCU--AAAUAAGAAGUGGUUACAGCA-AAUAAAAUUAUAAUUUAUUCGUUGC ....((((((((((--.....)))))))))).((((.-((((((.......)))))).)))). ( -18.10) >Bhalo.0 2799043 59 + 4202352 AAAAAGCCACUGUC-UAAAAAAGAAGUGGCUU--GUA-AUCAUCGUUCAAUUUCAAUCAGUUC ...((((((((.((-(.....)))))))))))--...-......................... ( -13.10) >Blich.0 185506 59 - 4222334 AAAAAGCCGCCCC----AAAAUGGAGCGGCCUUAGUAUGAUGAAAUCAUUAAGACACCGGCUC .....(((((.((----.....)).)))))....((.(((((....)))))..))........ ( -15.70) >consensus AAAAAACCACUUCU__AAAAAAGAAGUGGCUACAGCA_AAUAAAAUUAUAAUUUAAUCGGUGC ....((((((((((.......))))))))))................................ (-11.19 = -11.12 + -0.06)
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:27:42 2006