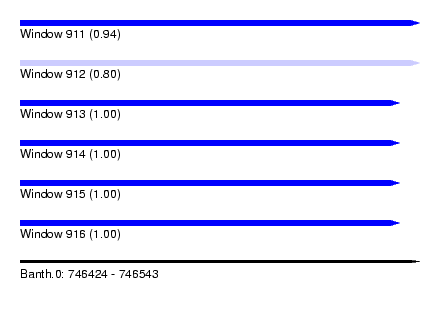
| Sequence ID | Banth.0 |
|---|---|
| Location | 746,424 – 746,543 |
| Length | 119 |
| Max. P | 0.998461 |

| Location | 746,424 – 746,543 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 53.49 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -20.93 |
| Energy contribution | -19.30 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 746424 119 + 5227293/0-120 CGGCGGUUCGAUCCCGUCAUCCUCCACCAUUCUUAAUUAAUGG-GCUAUAGCCAAGCGGUAAGGCAACGGACUUUGACUCCGUCAUGCGCUGGUUCGAAUCCAGCUAGCCCAGCCAUUAA .((((((..(((......)))....)))............(((-((((..(((....)))...((((((((.......)))))..)))(((((.......)))))))))))))))..... ( -37.50) >Bcere.0 823025 119 + 5224283/0-120 CGGCAGUUCGAAUCUGCCCCCCUCCACCAUUCUUAAUUAAUGG-GCUAUAGCCAAGCGGUAAGGCAACGGACUUUGACUCCGUCAUGCGCUGGUUCGAAUCCAGCUAGCCCAGCCAUUAA .(((((.......)))))...............((((...(((-((((..(((....)))...((((((((.......)))))..)))(((((.......))))))))))))...)))). ( -38.00) >Bhalo.0 1024949 116 + 4202352/0-120 CGCUGGUUCGAAUCCAGCUAGCCCAGCCAAUAAGCGGAAGUAGUUCAGUGGUAGAACACCACCUUGCCAAGGUGGGGGUCGCGGGUUCG---AAUCCCGUCUUCCGCUCCAAAGCUU-UG .(((((.......)))))......(((.....((((((((..((((.......)))).((((((.....)))))).....(((((....---...))))))))))))).....))).-.. ( -42.00) >Bclau.0 1447665 116 + 4303871/0-120 CCCUGGUUCGAAUCCAGGUAGCCCAGCCA-UAUGCGGAAGUAGUUCAGUGGUAGAAUAUCACCUUGCCAAGGUGGGGGUCGCGGGUUCG---AAUCCCGUCUUCCGCUUUAGUUUUUACG ....(((((((((((..(..((((.((((-(.(((....))).....)))))......((((((.....))))))))))..))))))))---))))........................ ( -38.90) >consensus CGCCGGUUCGAAUCCAGCUACCCCAACCAUUAUGAAGAAAUAG_GCAAUAGCAAAACAGCAACGCAACAAACUGGGACUCCCGCAUGCG___AAUCCAAUCCACCGAGCCAAGCCAUUAA .(((((.......)))))...............(((((..((((((((((((.....))))))))))))((((((((.................)))))))))))))............. (-20.93 = -19.30 + -1.63)

| Location | 746,424 – 746,543 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 53.49 |
| Mean single sequence MFE | -40.27 |
| Consensus MFE | -18.77 |
| Energy contribution | -15.40 |
| Covariance contribution | -3.37 |
| Combinations/Pair | 1.68 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 746424 119 + 5227293/0-120 UUAAUGGCUGGGCUAGCUGGAUUCGAACCAGCGCAUGACGGAGUCAAAGUCCGUUGCCUUACCGCUUGGCUAUAGC-CCAUUAAUUAAGAAUGGUGGAGGAUGACGGGAUCGAACCGCCG (((((...((((((((((((.......)))))....((((((.......))))))(((.........)))..))))-)))...)))))....(((((..(((......)))...))))). ( -39.80) >Bcere.0 823025 119 + 5224283/0-120 UUAAUGGCUGGGCUAGCUGGAUUCGAACCAGCGCAUGACGGAGUCAAAGUCCGUUGCCUUACCGCUUGGCUAUAGC-CCAUUAAUUAAGAAUGGUGGAGGGGGGCAGAUUCGAACUGCCG ...(((((..(((..(((((.......)))))(((..(((((.......))))))))......)))..)))))..(-((....((((....))))....)))(((((.......))))). ( -42.10) >Bhalo.0 1024949 116 + 4202352/0-120 CA-AAGCUUUGGAGCGGAAGACGGGAUU---CGAACCCGCGACCCCCACCUUGGCAAGGUGGUGUUCUACCACUGAACUACUUCCGCUUAUUGGCUGGGCUAGCUGGAUUCGAACCAGCG ..-.((((...(((((((((..(((..(---((......)))..)))....(((...(((((((...)))))))...))))))))))))...))))......(((((.......))))). ( -45.60) >Bclau.0 1447665 116 + 4303871/0-120 CGUAAAAACUAAAGCGGAAGACGGGAUU---CGAACCCGCGACCCCCACCUUGGCAAGGUGAUAUUCUACCACUGAACUACUUCCGCAUA-UGGCUGGGCUACCUGGAUUCGAACCAGGG .............(((((((..(((..(---((......)))..)))..........((((......)))).........)))))))...-((((...))))(((((.......))))). ( -33.60) >consensus CUAAAGGCUGGGAGAGCAAGACGCGAAC___CGAACCACCGACCCAAACCCCGGCAACGUACCGCUCGACCACAGA_CCACUAACGAAGAAUGGCGGAGCUAGCCGGAUUCGAACCACCG .............(((((((..(((..........)))..((((..((((((...))))))..)))).............)))))))...............(((((.......))))). (-18.77 = -15.40 + -3.37)


| Location | 746,424 – 746,537 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.18 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -36.20 |
| Energy contribution | -32.88 |
| Covariance contribution | -3.32 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 746424 113 + 5227293/200-320 GCUCACCAUUUUAACAAAAUAU--GCGGGUGUGGCGGAAUUGGCAGACGCACUAGACUUAGGAUCUAGCGCC---UUU--GGCGUGGGGGUUCGACUCCCUUCACCCGCACUUUUAAUAA ................(((..(--(((((((..........(.((((.((.((((((....).))))).)).---)))--).)..(((((.....)))))..))))))))..)))..... ( -36.50) >Bcere.0 823025 113 + 5224283/200-320 GCUCACCAUUUUAACAAAAUAU--GCGGGUGUGGCGGAAUUGGCAGACGCACUAGACUUAGGAUCUAGCGCC---UUU--GGCGUGGGGGUUCGACUCCCUUCACCCGCACUUUUAAUAA ................(((..(--(((((((..........(.((((.((.((((((....).))))).)).---)))--).)..(((((.....)))))..))))))))..)))..... ( -36.50) >Bsubt.0 95417 105 + 4214630/200-320 GCUCACCAUUUUA--------C--GCGGGUGUGGCGGAAUUGGCAGACGCGCUAGACUUAGGAUCUAGUGUC---UUUAUGACGUGGGGGUUCAAGUCCCUUCACCCGCAUUAU--AUAG .............--------.--(((((((.((.(((.(((((...((((((((((....).))))))(((---.....))))))...).)))).))))).))))))).....--.... ( -36.10) >Bhalo.0 1024949 114 + 4202352/200-320 AUCUUUCCACGUAAUUCCUU----GCCGGGGUGGUGGAAUUGGCAGACACACAGGACUUAAAAUCCUGCGGUAGGUGACUACCGUGCCGGUUCGAGUCCGGCCCUCGGCACCAU--UUUA ...................(----(((((((((((.(((((((((......(((((.......)))))((((((....))))))))))))))).)..)).)))).)))))....--.... ( -46.80) >Bclau.0 1447665 118 + 4303871/200-320 AAAAUAAGUUGUAGUCCUAGAUAGGCCGGGGUGGUGGAAUUGGCAGACACACAGGACUUAAAAUCCUGCGGUAGGUGACUACCGUGCCGGUUCGAGUCCGGCCCUCGGCAUCUU--AUGA ...(((((.(((.(.(((....))).)(((.((((.(((((((((......(((((.......)))))((((((....))))))))))))))).)..))).)))...))).)))--)).. ( -47.00) >consensus GCUCACCAUUUUAACAAAAUAU__GCGGGUGUGGCGGAAUUGGCAGACGCACUAGACUUAGGAUCUAGCGCC___UUU_UGCCGUGGGGGUUCGAGUCCCUUCACCCGCACUUU__AUAA ........................(((((((..(((...........))).(((((.......)))))((((((....)))))).(((((.......))))))))))))........... (-36.20 = -32.88 + -3.32)

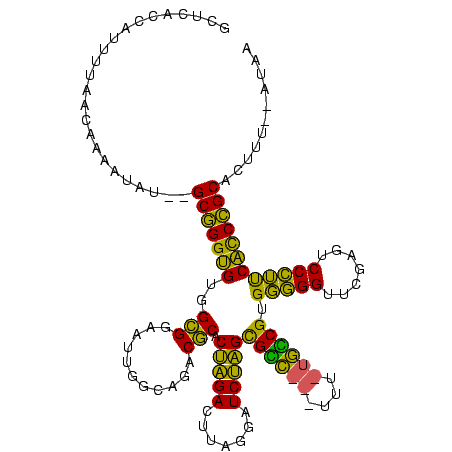
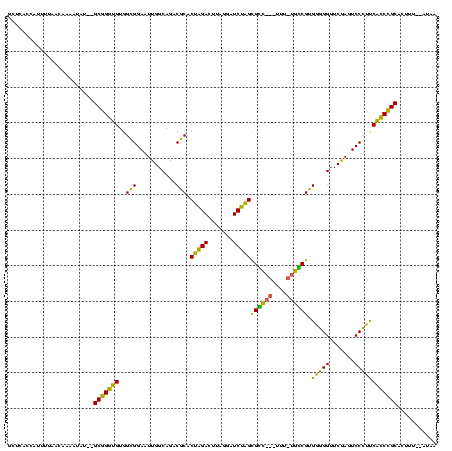
| Location | 746,424 – 746,537 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.18 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -28.48 |
| Energy contribution | -25.68 |
| Covariance contribution | -2.80 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 746424 113 + 5227293/200-320 UUAUUAAAAGUGCGGGUGAAGGGAGUCGAACCCCCACGCC--AAA---GGCGCUAGAUCCUAAGUCUAGUGCGUCUGCCAAUUCCGCCACACCCGC--AUAUUUUGUUAAAAUGGUGAGC (((.((((((((((((((..(((.(.....).)))..(.(--(.(---.(((((((((.....))))))))).).)).)..........)))))))--)).))))).))).......... ( -37.20) >Bcere.0 823025 113 + 5224283/200-320 UUAUUAAAAGUGCGGGUGAAGGGAGUCGAACCCCCACGCC--AAA---GGCGCUAGAUCCUAAGUCUAGUGCGUCUGCCAAUUCCGCCACACCCGC--AUAUUUUGUUAAAAUGGUGAGC (((.((((((((((((((..(((.(.....).)))..(.(--(.(---.(((((((((.....))))))))).).)).)..........)))))))--)).))))).))).......... ( -37.20) >Bsubt.0 95417 105 + 4214630/200-320 CUAU--AUAAUGCGGGUGAAGGGACUUGAACCCCCACGUCAUAAA---GACACUAGAUCCUAAGUCUAGCGCGUCUGCCAAUUCCGCCACACCCGC--G--------UAAAAUGGUGAGC ((((--.(.(((((((((..(((((.((......)).)))....(---(((.((((((.....))))))...)))).......))....)))))))--)--------).).))))..... ( -30.90) >Bhalo.0 1024949 114 + 4202352/200-320 UAAA--AUGGUGCCGAGGGCCGGACUCGAACCGGCACGGUAGUCACCUACCGCAGGAUUUUAAGUCCUGUGUGUCUGCCAAUUCCACCACCCCGGC----AAGGAAUUACGUGGAAAGAU ....--....(((((.(((((((.......)))))..(((((.(((.....(((((((.....)))))))))).)))))..........)).))))----)................... ( -41.40) >Bclau.0 1447665 118 + 4303871/200-320 UCAU--AAGAUGCCGAGGGCCGGACUCGAACCGGCACGGUAGUCACCUACCGCAGGAUUUUAAGUCCUGUGUGUCUGCCAAUUCCACCACCCCGGCCUAUCUAGGACUACAACUUAUUUU ....--.((((((((.(((((((.......)))))..(((((.(((.....(((((((.....)))))))))).)))))..........)).))))..)))).................. ( -41.60) >consensus UUAU__AAAGUGCGGGUGAAGGGACUCGAACCCCCACGCCA_AAA___GCCGCUAGAUCCUAAGUCUAGUGCGUCUGCCAAUUCCGCCACACCCGC__AUAUUGUAUUAAAAUGGUGAGC ...........(((((((..(((.......)))....(((((....)))))(((((((.....)))))))(((...........)))..)))))))........................ (-28.48 = -25.68 + -2.80)

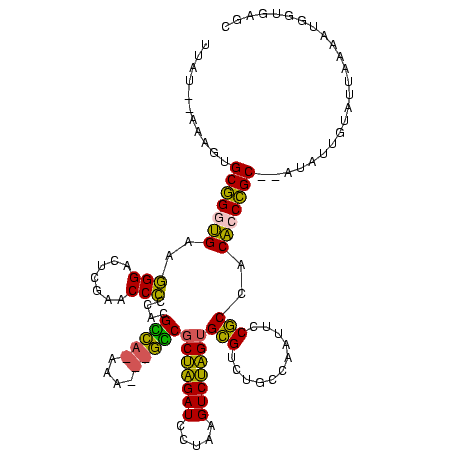
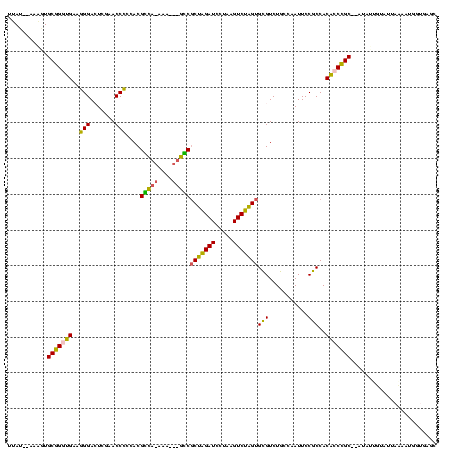
| Location | 746,424 – 746,537 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.12 |
| Mean single sequence MFE | -40.94 |
| Consensus MFE | -36.20 |
| Energy contribution | -32.88 |
| Covariance contribution | -3.32 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 746424 113 + 5227293/210-330 UUAACAAAAUAU--GCGGGUGUGGCGGAAUUGGCAGACGCACUAGACUUAGGAUCUAGCGCC---UUU--GGCGUGGGGGUUCGACUCCCUUCACCCGCACUUUUAAUAAAAUAACUCAU ......(((..(--(((((((..........(.((((.((.((((((....).))))).)).---)))--).)..(((((.....)))))..))))))))..)))............... ( -36.50) >Bcere.0 823025 113 + 5224283/210-330 UUAACAAAAUAU--GCGGGUGUGGCGGAAUUGGCAGACGCACUAGACUUAGGAUCUAGCGCC---UUU--GGCGUGGGGGUUCGACUCCCUUCACCCGCACUUUUAAUAAAAUAACUCAU ......(((..(--(((((((..........(.((((.((.((((((....).))))).)).---)))--).)..(((((.....)))))..))))))))..)))............... ( -36.50) >Bsubt.0 95417 105 + 4214630/210-330 UUA--------C--GCGGGUGUGGCGGAAUUGGCAGACGCGCUAGACUUAGGAUCUAGUGUC---UUUAUGACGUGGGGGUUCAAGUCCCUUCACCCGCAUUAU--AUAGGAUAACAGUU ...--------.--(((((((.((.(((.(((((...((((((((((....).))))))(((---.....))))))...).)))).))))).))))))).....--.............. ( -36.10) >Bhalo.0 1024949 114 + 4202352/210-330 GUAAUUCCUU----GCCGGGGUGGUGGAAUUGGCAGACACACAGGACUUAAAAUCCUGCGGUAGGUGACUACCGUGCCGGUUCGAGUCCGGCCCUCGGCACCAU--UUUAGACAAGCUUU .......(((----((..(((((((((((((((((......(((((.......)))))((((((....)))))))))))))))....(((.....)))))))))--))..).)))).... ( -47.00) >Bclau.0 1447665 118 + 4303871/210-330 GUAGUCCUAGAUAGGCCGGGGUGGUGGAAUUGGCAGACACACAGGACUUAAAAUCCUGCGGUAGGUGACUACCGUGCCGGUUCGAGUCCGGCCCUCGGCAUCUU--AUGAAACGACAUGA ...(((........(((((((((((.(((((((((......(((((.......)))))((((((....))))))))))))))).)..)).)))).)))).((..--..))...))).... ( -48.60) >consensus UUAACAAAAUAU__GCGGGUGUGGCGGAAUUGGCAGACGCACUAGACUUAGGAUCUAGCGCC___UUU_UGCCGUGGGGGUUCGAGUCCCUUCACCCGCACUUU__AUAAAAUAACACAU ..............(((((((..(((...........))).(((((.......)))))((((((....)))))).(((((.......))))))))))))..................... (-36.20 = -32.88 + -3.32)

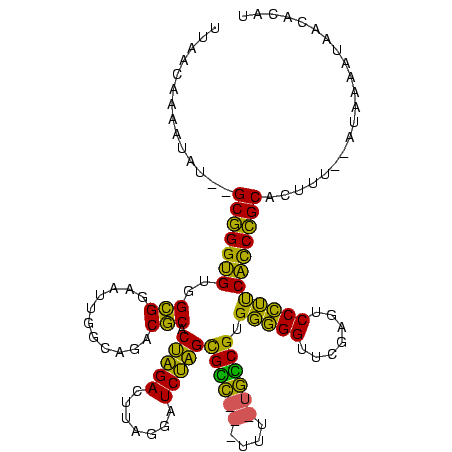

| Location | 746,424 – 746,537 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.12 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -28.48 |
| Energy contribution | -25.68 |
| Covariance contribution | -2.80 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.76 |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.998461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 746424 113 + 5227293/210-330 AUGAGUUAUUUUAUUAAAAGUGCGGGUGAAGGGAGUCGAACCCCCACGCC--AAA---GGCGCUAGAUCCUAAGUCUAGUGCGUCUGCCAAUUCCGCCACACCCGC--AUAUUUUGUUAA ..............((((((((((((((..(((.(.....).)))..(.(--(.(---.(((((((((.....))))))))).).)).)..........)))))))--)).))))).... ( -36.70) >Bcere.0 823025 113 + 5224283/210-330 AUGAGUUAUUUUAUUAAAAGUGCGGGUGAAGGGAGUCGAACCCCCACGCC--AAA---GGCGCUAGAUCCUAAGUCUAGUGCGUCUGCCAAUUCCGCCACACCCGC--AUAUUUUGUUAA ..............((((((((((((((..(((.(.....).)))..(.(--(.(---.(((((((((.....))))))))).).)).)..........)))))))--)).))))).... ( -36.70) >Bsubt.0 95417 105 + 4214630/210-330 AACUGUUAUCCUAU--AUAAUGCGGGUGAAGGGACUUGAACCCCCACGUCAUAAA---GACACUAGAUCCUAAGUCUAGCGCGUCUGCCAAUUCCGCCACACCCGC--G--------UAA ..............--...(((((((((..(((((.((......)).)))....(---(((.((((((.....))))))...)))).......))....)))))))--)--------).. ( -30.00) >Bhalo.0 1024949 114 + 4202352/210-330 AAAGCUUGUCUAAA--AUGGUGCCGAGGGCCGGACUCGAACCGGCACGGUAGUCACCUACCGCAGGAUUUUAAGUCCUGUGUGUCUGCCAAUUCCACCACCCCGGC----AAGGAAUUAC ....((((((....--.(((((..(((.(((((.......)))))..(((((.(((.....(((((((.....)))))))))).)))))..))))))))....)))----)))....... ( -42.40) >Bclau.0 1447665 118 + 4303871/210-330 UCAUGUCGUUUCAU--AAGAUGCCGAGGGCCGGACUCGAACCGGCACGGUAGUCACCUACCGCAGGAUUUUAAGUCCUGUGUGUCUGCCAAUUCCACCACCCCGGCCUAUCUAGGACUAC ....(((.(.....--.((((((((.(((((((.......)))))..(((((.(((.....(((((((.....)))))))))).)))))..........)).))))..))))).)))... ( -42.50) >consensus AAAAGUUAUUUUAU__AAAGUGCGGGUGAAGGGACUCGAACCCCCACGCCA_AAA___GCCGCUAGAUCCUAAGUCUAGUGCGUCUGCCAAUUCCGCCACACCCGC__AUAUUGUAUUAA .....................(((((((..(((.......)))....(((((....)))))(((((((.....)))))))(((...........)))..))))))).............. (-28.48 = -25.68 + -2.80)

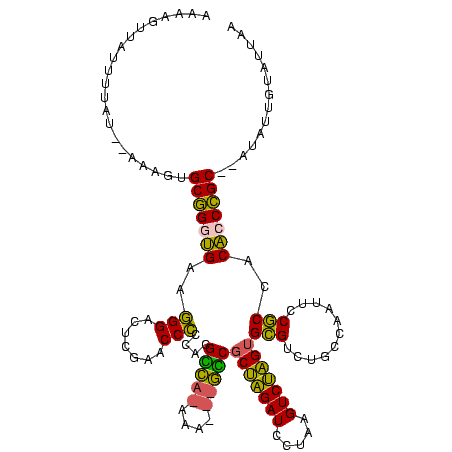
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:27:32 2006