| Sequence ID | Banth.0 |
|---|---|
| Location | 741,570 – 741,690 |
| Length | 120 |
| Max. P | 0.998940 |

| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -38.63 |
| Consensus MFE | -37.66 |
| Energy contribution | -36.23 |
| Covariance contribution | -1.43 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
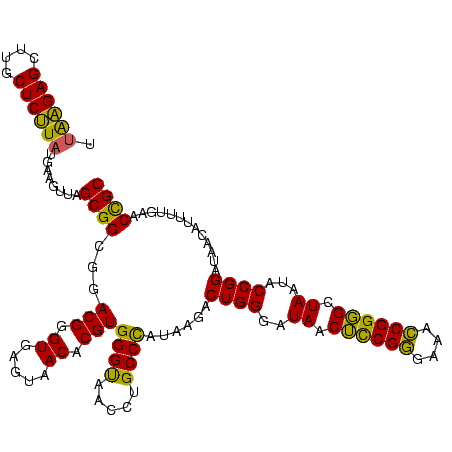
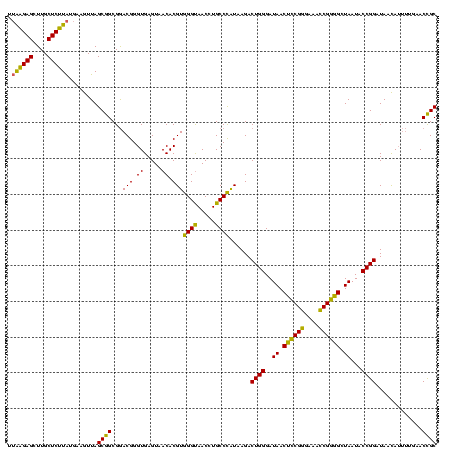
>Banth.0 741570 120 + 5227293/40-160 UUAAGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGC .((((((....))))))...........((((..((((..((.((......)).))..))))......((((..((.((((((....)))))).))...)))).............)))) ( -40.50) >Bcere.0 818171 120 + 5224283/40-160 UUAAGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGC .((((((....))))))...........((((..((((..((.((......)).))..))))......((((..((.((((((....)))))).))...)))).............)))) ( -40.50) >Bhalo.0 50621 118 + 4202352/40-160 A-GGGAGCUUGCUCCUG-GAGGUUAGCGGCGAACGGGUGAGUAACACGUGGGCAACCUGCCUGUAAGACUGGGAUAACAUCGAGAAAUCGGUGCUAAUACCGGAUAAUAAAAAGAACUGC .-(((((....))))).-..((((.((((((.(((.((.....)).)))((....)))))).))....((((..((.((((((....)))))).))...))))...........)))).. ( -34.90) >consensus UUAAGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGC .((((((....))))))........((((...(((.((.....)).)))((((.....))))......((((..((.((((((....)))))).))...)))).............)))) (-37.66 = -36.23 + -1.43)

| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -45.53 |
| Consensus MFE | -47.73 |
| Energy contribution | -45.53 |
| Covariance contribution | -2.20 |
| Combinations/Pair | 1.38 |
| Mean z-score | -3.98 |
| Structure conservation index | 1.05 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
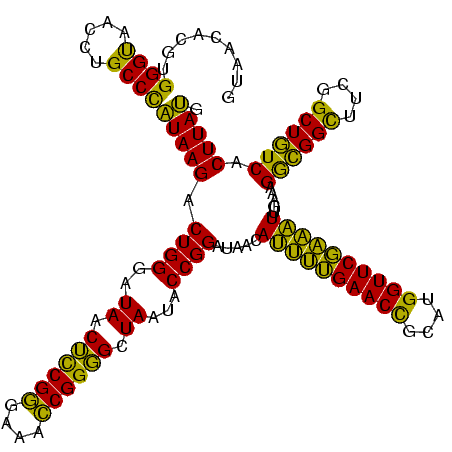
>Banth.0 741570 120 + 5227293/80-200 GUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUCGGCUGUCACUUAUG .........((((.....))))(((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((....)))))).))))). ( -49.10) >Bcere.0 818171 120 + 5224283/80-200 GUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUCGGCUGUCACUUAUG .........((((.....))))(((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((....)))))).))))). ( -49.10) >Bhalo.0 50621 120 + 4202352/80-200 GUAACACGUGGGCAACCUGCCUGUAAGACUGGGAUAACAUCGAGAAAUCGGUGCUAAUACCGGAUAAUAAAAAGAACUGCAUGGUUCUUUUUUGAAAGAUGGUUUCGGCUAUCACUUACA .........((((.....))))(((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((....)))))).))))). ( -38.40) >consensus GUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUCGGCUGUCACUUAUG .........((((.....))))(((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((....)))))).))))). (-47.73 = -45.53 + -2.20)

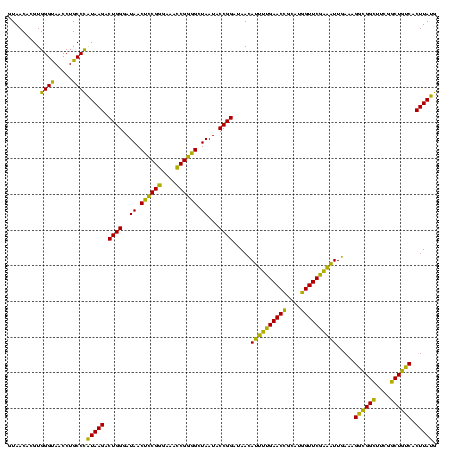
| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -39.87 |
| Consensus MFE | -35.81 |
| Energy contribution | -34.93 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/120-240 CGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUCGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGG (((....)))((((((...(((......((((((((((....)))))))))).....((((((....)))))).....))).))).))).(((..(.(((.(.....).))).)..))). ( -43.10) >Bcere.0 818171 120 + 5224283/120-240 CGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUCGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGG (((....)))((((((...(((......((((((((((....)))))))))).....((((((....)))))).....))).))).))).(((..(.(((.(.....).))).)..))). ( -43.10) >Bhalo.0 50621 120 + 4202352/120-240 CGAGAAAUCGGUGCUAAUACCGGAUAAUAAAAAGAACUGCAUGGUUCUUUUUUGAAAGAUGGUUUCGGCUAUCACUUACAGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUGGGGUAACGG (((....))).((((..(((((......((((((((((....)))))))))).....((((((....))))))(((..(.((((.(((...))).)))).)..)))))))).)))).... ( -33.40) >consensus CGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUCGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGG (((....))).......((((.......((((((((((....)))))))))).....((((((....))))))............(((.((...((....))..)).)))..)))).... (-35.81 = -34.93 + -0.87)

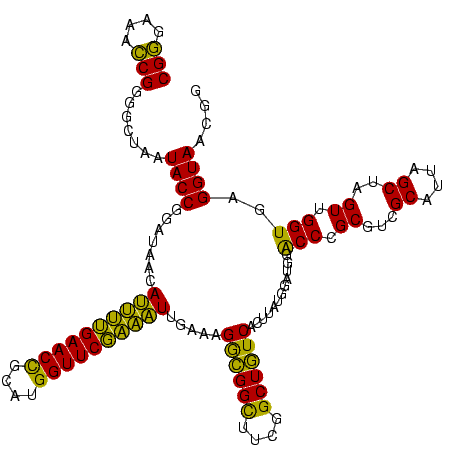
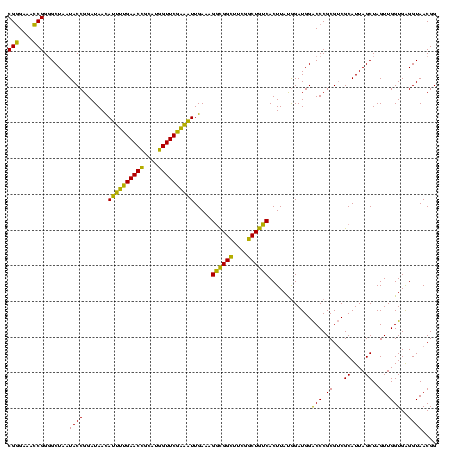
| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -45.67 |
| Consensus MFE | -41.12 |
| Energy contribution | -44.30 |
| Covariance contribution | 3.18 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/160-280 AUGGUUCGAAAUUGAAAGGCGGCUUCGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGA ..((((((....(((..((((((....))))))..)))....))))))(((((((.....(((..((((((((.......)))))))))))..)))))))......((((....)))).. ( -46.80) >Bcere.0 818171 120 + 5224283/160-280 AUGGUUCGAAAUUGAAAGGCGGCUUCGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGA ..((((((....(((..((((((....))))))..)))....))))))(((((((.....(((..((((((((.......)))))))))))..)))))))......((((....)))).. ( -46.80) >Bhalo.0 50621 120 + 4202352/160-280 AUGGUUCUUUUUUGAAAGAUGGUUUCGGCUAUCACUUACAGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUGGGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGA ..(((((.(((.(((..((((((....))))))..))).))).))))).((.(((((((.(((..((((((((.......)))))))))))...))))))).))..((((....)))).. ( -43.40) >consensus AUGGUUCGAAAUUGAAAGGCGGCUUCGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGA ..(((((.....(((..((((((....))))))..))).....))))).((..((((((.(((..((((((((.......)))))))))))...))))))..))..((((....)))).. (-41.12 = -44.30 + 3.18)

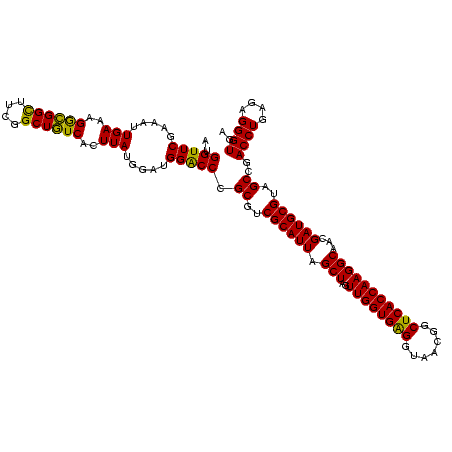
| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -48.70 |
| Consensus MFE | -47.32 |
| Energy contribution | -47.77 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/200-320 CGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUC ..........(((((((((((.((......((((((((((((.........))))))))))))...((((..((((((((.......)))))))).))))...)).))))).)))))).. ( -50.20) >Bcere.0 818171 120 + 5224283/200-320 CGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUC ..........(((((((((((.((......((((((((((((.........))))))))))))...((((..((((((((.......)))))))).))))...)).))))).)))))).. ( -50.20) >Bhalo.0 50621 120 + 4202352/200-320 CGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCCCACCAACUAGCUAAUGCGCCGCGGGCCCAUC ..........((((((..............((((((((((((.........))))))))))))..(((.((.((((((...........))))))...((....)))).))))))))).. ( -45.70) >consensus CGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUC ..........(((((((((((.((......((((((((((((.........))))))))))))......((.((((((((.......))))))))...))...)).))))).)))))).. (-47.32 = -47.77 + 0.45)

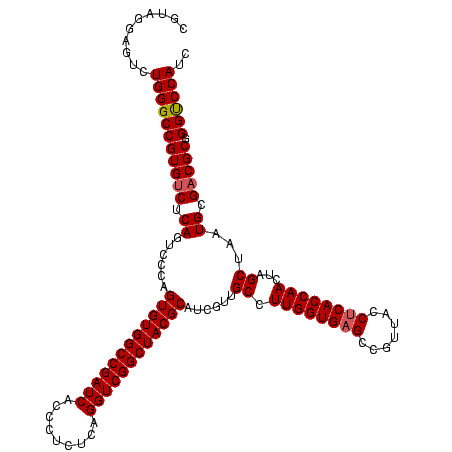
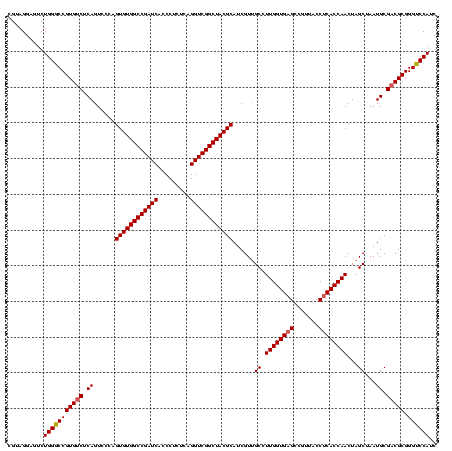
| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -48.13 |
| Consensus MFE | -43.13 |
| Energy contribution | -48.13 |
| Covariance contribution | 5.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.689233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/240-360 CUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUC ....(((..((.((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...))..(((....)))))..)))((....)). ( -48.10) >Bcere.0 818171 120 + 5224283/240-360 CUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUC ....(((..((.((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...))..(((....)))))..)))((....)). ( -48.10) >Bhalo.0 50621 120 + 4202352/240-360 CUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUC ....(((..((.((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...))..(((....)))))..)))((....)). ( -48.20) >consensus CUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUC ....(((..((.((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...))..(((....)))))..)))((....)). (-43.13 = -48.13 + 5.00)

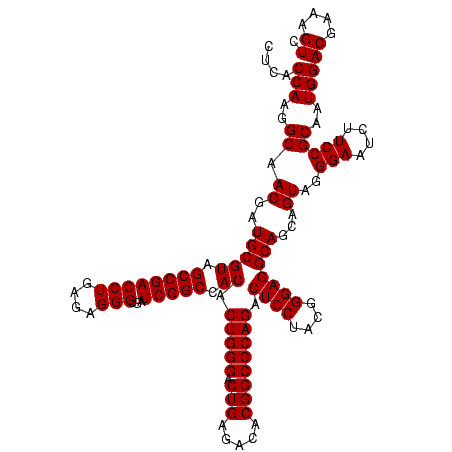
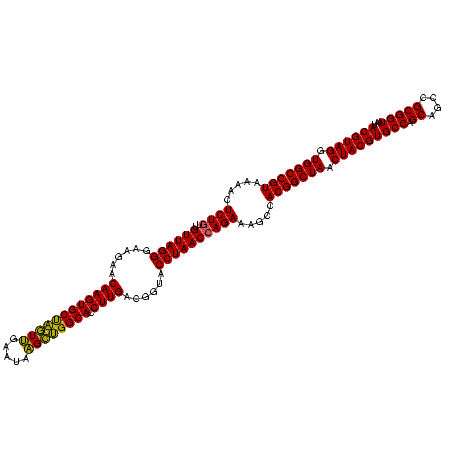
| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -42.73 |
| Consensus MFE | -39.30 |
| Energy contribution | -40.87 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/360-480 UGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACG ...((......((.(((....))).))..(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..))))))).)) ( -41.90) >Bcere.0 818171 120 + 5224283/360-480 UGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACG ...((......((.(((....))).))..(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..))))))).)) ( -41.90) >Bhalo.0 50621 119 + 4202352/360-480 UGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAA-AGGGCGGCACCUUGACGGUACCUAACGAGAAAGCCACG ...((......((.(((....))).))..((((((((((.........))).((((((((......((((((((((((...-.)))))))).))))......))))))))))))))).)) ( -44.40) >consensus UGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACG ...((......((.(((....))).))..(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..))))))).)) (-39.30 = -40.87 + 1.56)

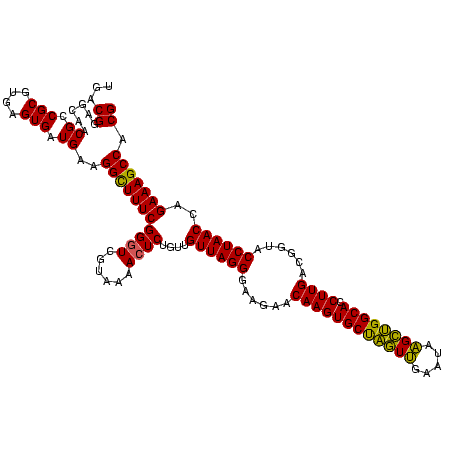
| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -44.07 |
| Consensus MFE | -43.45 |
| Energy contribution | -42.23 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/400-520 CGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGC (((.....((((..((((((......((((((((((((.....)))))))).))))......)))))))))).....)))((((.(((((((((.((....)))))...)))))).)))) ( -42.20) >Bcere.0 818171 120 + 5224283/400-520 CGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGC (((.....((((..((((((......((((((((((((.....)))))))).))))......)))))))))).....)))((((.(((((((((.((....)))))...)))))).)))) ( -42.20) >Bhalo.0 50621 119 + 4202352/400-520 CGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAA-AGGGCGGCACCUUGACGGUACCUAACGAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGC (((....((.(.((((((((......((((((((((((...-.)))))))).))))......)))))))).).))..)))((((.(((((((((.((....)))))...)))))).)))) ( -47.80) >consensus CGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGC (((.....((((..((((((......((((((((((((.....)))))))).))))......)))))))))).....)))((((.(((((((((.((....)))))...)))))).)))) (-43.45 = -42.23 + -1.22)

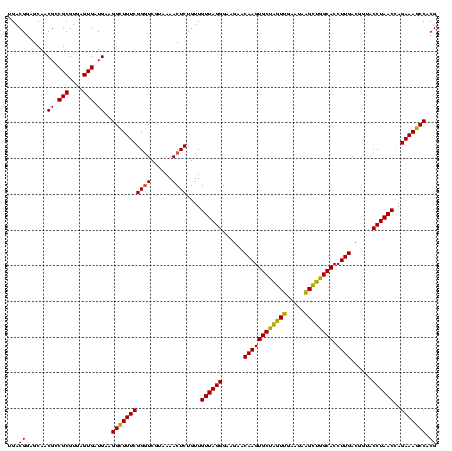
| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -26.51 |
| Consensus MFE | -23.90 |
| Energy contribution | -23.35 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/560-680 CUCUACGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAGAAACCA .........((((...((.......((((.......))))......))....((((((....(((......)))..(((((((.....)))))))..........))))))..))))... ( -24.02) >Bcere.0 818171 120 + 5224283/560-680 CUCUACGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAGAAACCA .........((((...((.......((((.......))))......))....((((((....(((......)))..(((((((.....)))))))..........))))))..))))... ( -24.02) >Bhalo.0 50621 120 + 4202352/560-680 CUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGUACUCAAGUCUCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAGAGACCG ...(((((........)).....((((....))))...........))).....(((((..(((((....(((...(((((((.....)))))))...)))....)))))...))))).. ( -31.50) >consensus CUCUACGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAGAAACCA .........((((...((.......((((.......))))......))....((((((....(((......)))..(((((((.....)))))))..........))))))..))))... (-23.90 = -23.35 + -0.55)

| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -36.26 |
| Energy contribution | -35.60 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.28 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/600-720 UGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCU (.(((..((...(....)...))..))).)...(((((.......(((...((((((((.(.(((......)))....).))))))))...))).(((.((....)).)))))))).... ( -34.30) >Bcere.0 818171 120 + 5224283/600-720 UGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCU (.(((..((...(....)...))..))).)...(((((.......(((...((((((((.(.(((......)))....).))))))))...))).(((.((....)).)))))))).... ( -34.30) >Bhalo.0 50621 120 + 4202352/600-720 GGGAGGGUCAUUGGAAACUGGGAGACUUGAGUACAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCU .((((((((.......(((((....)...................(((...((((((((.(.(((......)))....).))))))))...))))))).((....))))))))))..... ( -38.40) >consensus UGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCU .((((((((.......(((((....)...................(((...((((((((.(.(((......)))....).))))))))...))))))).((....))))))))))..... (-36.26 = -35.60 + -0.66)


| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -40.13 |
| Consensus MFE | -38.96 |
| Energy contribution | -38.30 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/640-760 GGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAG .....(((...((((((((.(.(((......)))....).))))))))...)))(((((((....))((((....)))).........)))))..((((....))))............. ( -38.20) >Bcere.0 818171 120 + 5224283/640-760 GGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAG .....(((...((((((((.(.(((......)))....).))))))))...)))(((((((....))((((....)))).........)))))..((((....))))............. ( -38.20) >Bhalo.0 50621 120 + 4202352/640-760 GGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAG ((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).))))))).((.(((..((.((((....))((....)))).))..))).)).. ( -44.00) >consensus GGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAG .....(((...((((((((.(.(((......)))....).))))))))...)))(((((((....))((((....)))).........)))))..((((....))))............. (-38.96 = -38.30 + -0.66)

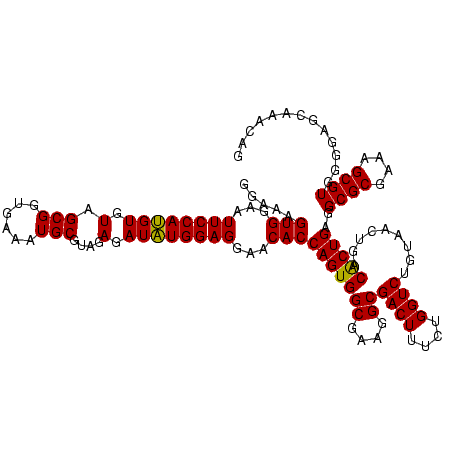
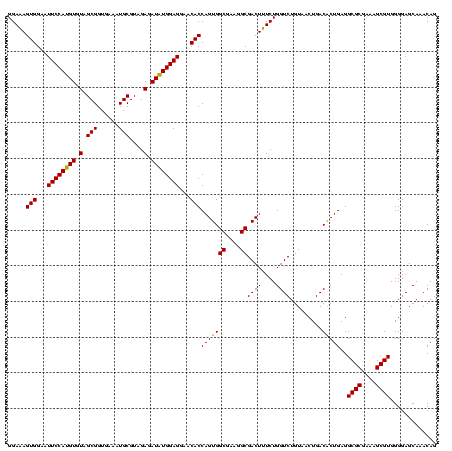
| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -41.97 |
| Consensus MFE | -43.38 |
| Energy contribution | -41.83 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.87 |
| Structure conservation index | 1.03 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/720-840 GUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUA .(((((..((((.((((((....))(((((..((...((((.........))))))..))))))))(....)..).))))...((..(((((((((....)))))))))..))..))))) ( -41.60) >Bcere.0 818171 120 + 5224283/720-840 GUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUA .(((((..((((.((((((....))(((((..((...((((.........))))))..))))))))(....)..).))))...((..(((((((((....)))))))))..))..))))) ( -41.60) >Bhalo.0 50621 120 + 4202352/720-840 GUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGACGCCCUUAGUGCCGAAGUUA .(((((..((((.((((((....))(((((..((...((((.........))))))..))))))))(....)..).))))...((..((((((((......))))))))..))..))))) ( -42.70) >consensus GUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUA .(((((..((((.((((((....))(((((..((...((((.........))))))..))))))))(....)..).))))...((..((((((((......))))))))..))..))))) (-43.38 = -41.83 + -1.55)

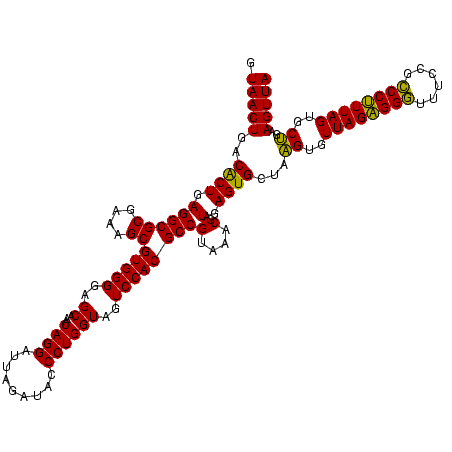
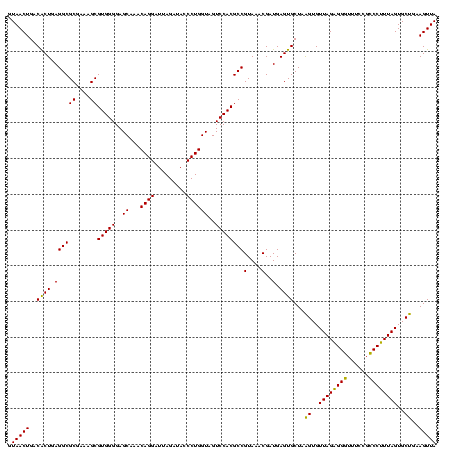
| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -41.57 |
| Consensus MFE | -43.43 |
| Energy contribution | -41.43 |
| Covariance contribution | -1.99 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.16 |
| Structure conservation index | 1.04 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/760-880 GAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGC .........(((.(((.........((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))).))).......(((....))) ( -41.60) >Bcere.0 818171 120 + 5224283/760-880 GAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGC .........(((.(((.........((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))).))).......(((....))) ( -41.60) >Bhalo.0 50621 120 + 4202352/760-880 GAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGACGCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGU .........(((.(((.........((....))..(((((((.((..((((((((......))))))))..))...((....))....))))))).))).))).......(((....))) ( -41.50) >consensus GAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGC .........(((.(((.........((....))..(((((((.((..((((((((......))))))))..))...((....))....))))))).))).))).......(((....))) (-43.43 = -41.43 + -1.99)

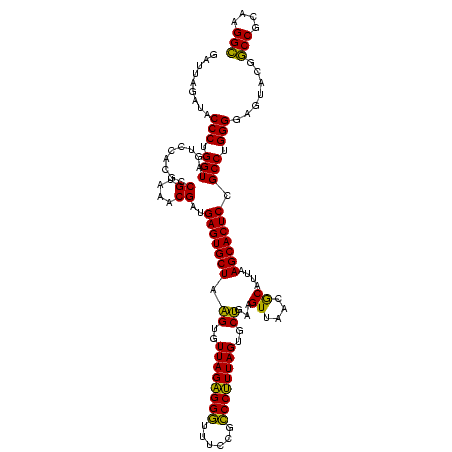
| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -47.20 |
| Consensus MFE | -47.44 |
| Energy contribution | -46.00 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.32 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/800-920 CUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUG ...((..(((((((((....)))))))))..))................((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).. ( -48.20) >Bcere.0 818171 120 + 5224283/800-920 CUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUG ...((..(((((((((....)))))))))..))................((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).. ( -48.20) >Bhalo.0 50621 120 + 4202352/800-920 CUAGGUGUUAGGGGUUUCGACGCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCAGUG ...((..((((((((......))))))))..))........(((.....((.((((((((...((((.(((((....)))))..))))..)))......)))))))..((....)).))) ( -45.20) >consensus CUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUG ...((..((((((((......))))))))..))................((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).. (-47.44 = -46.00 + -1.44)

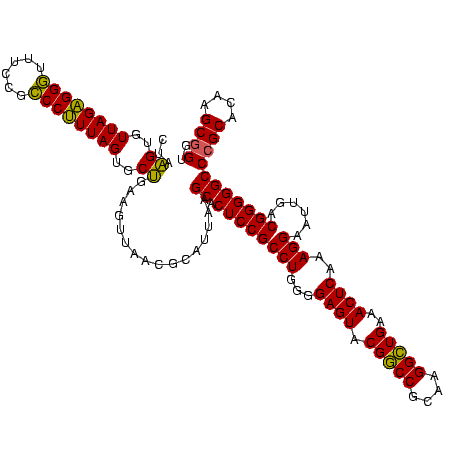
| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -40.43 |
| Consensus MFE | -39.83 |
| Energy contribution | -39.17 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/840-960 ACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUAC ..((.....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))....))....(((((..((((........))))))))).... ( -41.80) >Bcere.0 818171 120 + 5224283/840-960 ACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUAC ..((.....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))....))....(((((..((((........))))))))).... ( -41.80) >Bhalo.0 50621 120 + 4202352/840-960 ACACAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCAGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUAC ..((((......((((((((...((((.(((((....)))))..))))..)))......)))))((((((.......)))).))))))(((((..((((........))))))))).... ( -37.70) >consensus ACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUAC ..((((......((((((((...((((.(((((....)))))..))))..)))......)))))((((((.......)))).))))))(((((..((((........))))))))).... (-39.83 = -39.17 + -0.66)

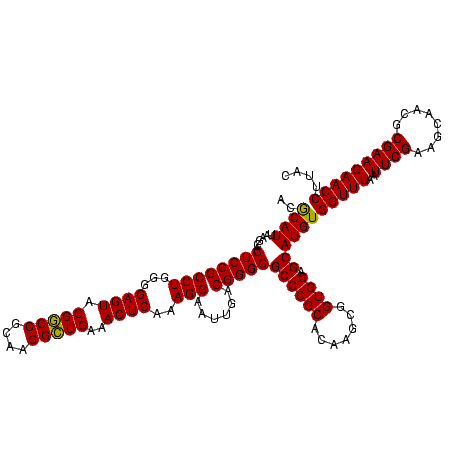
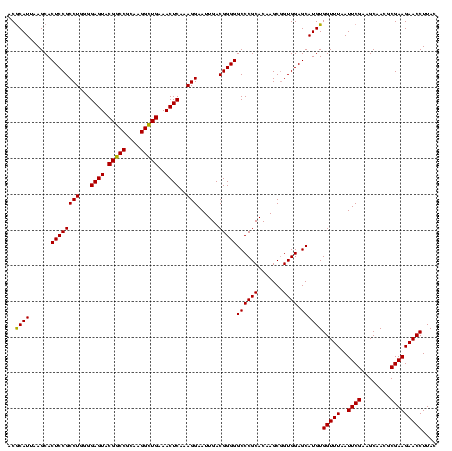
| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -36.37 |
| Consensus MFE | -37.46 |
| Energy contribution | -35.47 |
| Covariance contribution | -1.99 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.92 |
| Structure conservation index | 1.03 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/1200-1320 UACACACGUGCUACAAUGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCG .................(((((((.....(((..((...(((....)))...))....))).....)))))))((((((.....(((((((........)))))))...))))))..... ( -33.80) >Bcere.0 818171 120 + 5224283/1200-1320 UACACACGUGCUACAAUGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCG .................(((((((.....(((..((...(((....)))...))....))).....)))))))((((((.....(((((((........)))))))...))))))..... ( -33.80) >Bhalo.0 50621 120 + 4202352/1200-1320 UACACACGUGCUACAAUGGAUGGUACAAAGGGUUGCGAAGCCGCGAGGUGAAGCCAAUCCCAGAAAGCCAUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAAGCCGGAAUUG .............(((((((((((.....(((((((...(((....)))...).))))))......)))))))....(((((..(((((((........))))))).....))))))))) ( -41.50) >consensus UACACACGUGCUACAAUGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCG ......((.(((.((..(((((((.....(((..((...(((....)))...))....))).....)))))))...........(((((((........))))))))).)))))...... (-37.46 = -35.47 + -1.99)

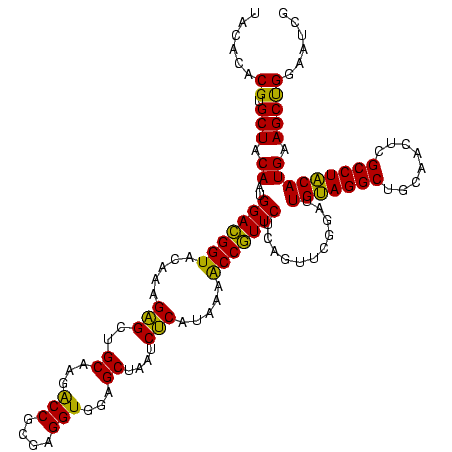
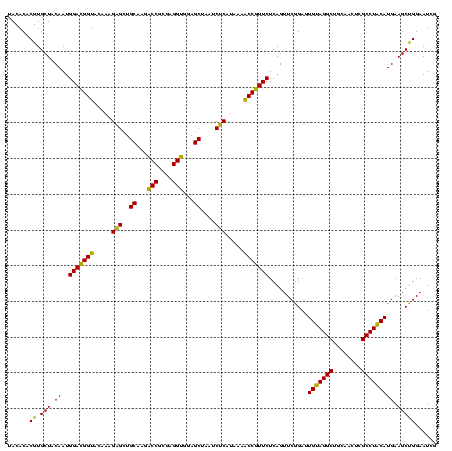
| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -45.30 |
| Consensus MFE | -44.57 |
| Energy contribution | -44.57 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/1360-1480 CGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACA (((((..((....))..)))))((((.((.(((..((((((..((((...((.((((..(((..((.....)).))).)))).))..))))..))))))..))))))))).......... ( -45.30) >Bcere.0 818171 119 + 5224283/1360-1480 CGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGGGGUAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACA (((((..((....))..)))))((((.((.(((..((((((..((((...((.((((..(((..((....-)).))).)))).))..))))..))))))..))))))))).......... ( -45.30) >Bhalo.0 50621 119 + 4202352/1360-1480 CGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGGGGUAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACA (((((..((....))..)))))((((.((.(((..((((((..((((...((.((((..(((..((....-)).))).)))).))..))))..))))))..))))))))).......... ( -45.30) >consensus CGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGGGGUAACCUUUU_GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACA (((((..((....))..)))))((((.((.(((..((((((..((((...((.((((..(((..((.....)).))).)))).))..))))..))))))..))))))))).......... (-44.57 = -44.57 + 0.00)


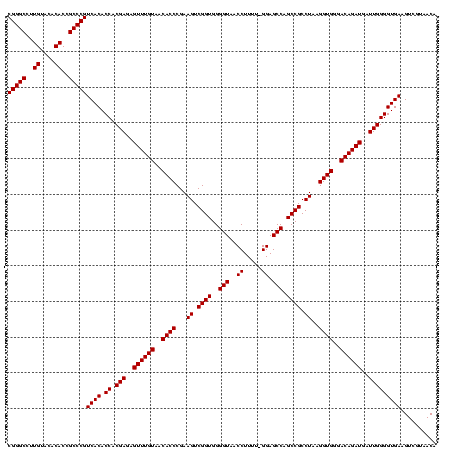
| Location | 741,570 – 741,690 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -45.30 |
| Consensus MFE | -44.57 |
| Energy contribution | -44.57 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 120 + 5227293/1400-1520 UAACACCCGAAGUCGGUGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUU .....((((.....((((((((..((.....)).)))..))))).....))))..........(((((((.((....))....((((((((((....))))))))))..))))))).... ( -45.30) >Bcere.0 818171 119 + 5224283/1400-1520 UAACACCCGAAGUCGGUGGGGUAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUU .....((((.....((((((((..((....-)).)))..))))).....))))..........(((((((.((....))....((((((((((....))))))))))..))))))).... ( -45.30) >Bhalo.0 50621 119 + 4202352/1400-1520 UAACACCCGAAGUCGGUGGGGUAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUU .....((((.....((((((((..((....-)).)))..))))).....))))..........(((((((.((....))....((((((((((....))))))))))..))))))).... ( -45.30) >consensus UAACACCCGAAGUCGGUGGGGUAACCUUUU_GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUU .....((((.....((((((((..((.....)).)))..))))).....))))..........(((((((.((....))....((((((((((....))))))))))..))))))).... (-44.57 = -44.57 + 0.00)

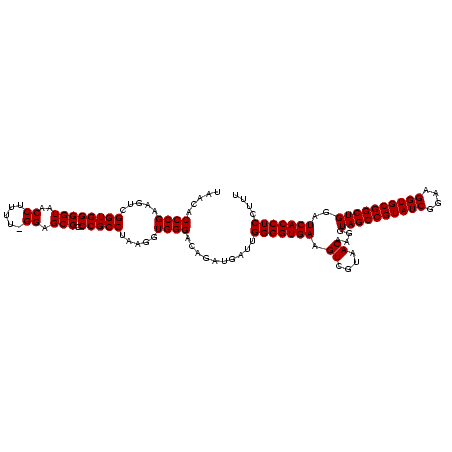
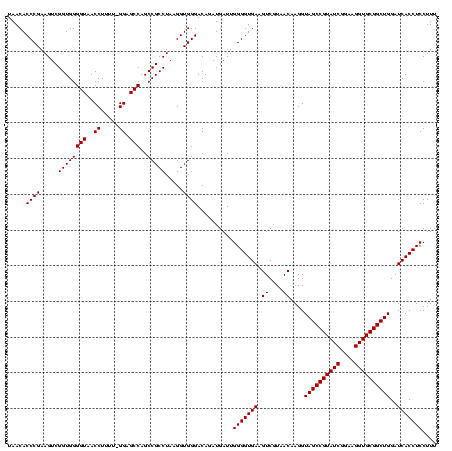
| Location | 741,570 – 741,680 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -41.93 |
| Consensus MFE | -34.03 |
| Energy contribution | -34.70 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 110 + 5227293/1440-1560 CGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA----UAAGCG------CUGCUUAUCA ((((...))))...((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))......(((----(((((.------..)))))))) ( -41.30) >Bcere.0 818171 110 + 5224283/1440-1560 CGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA----UGAACG------CUGUUCAUCA ((((...))))...((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))......(((----(((((.------..)))))))) ( -41.20) >Bhalo.0 50621 120 + 4202352/1440-1560 CGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUACGGAGUUUUUAACUCUAGUCGGACCAUUCGCAGAUUC .((....(((((..(.(((((..(((((((.((....))....((((((((((....))))))))))..))))).))..))...(((((.....))))).))))..))))).))...... ( -43.30) >consensus CGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA____UAAACG______CUGCUCAUCA ((((...))))...((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..)).............((((((.......))))))... (-34.03 = -34.70 + 0.67)

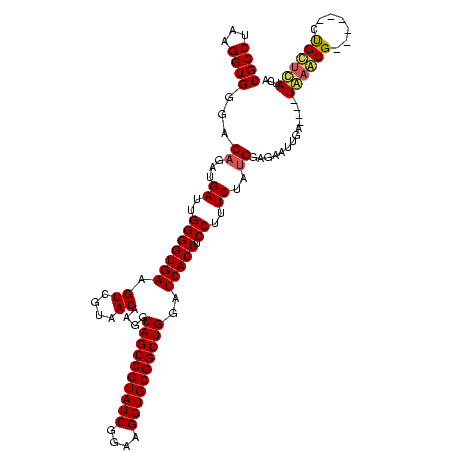
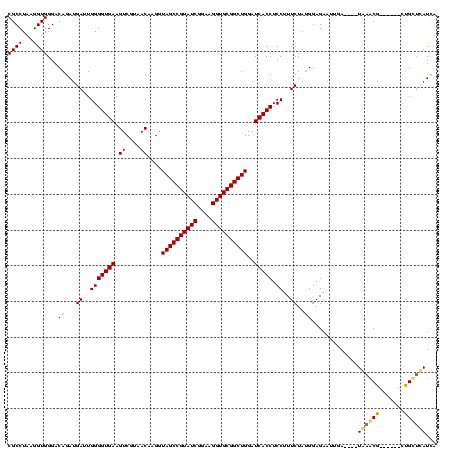
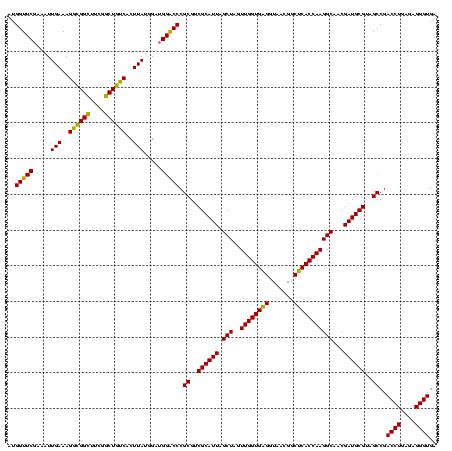
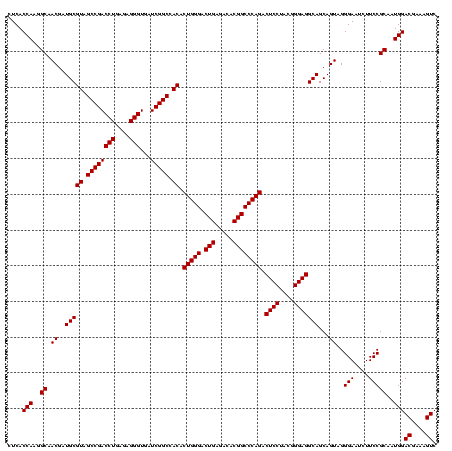
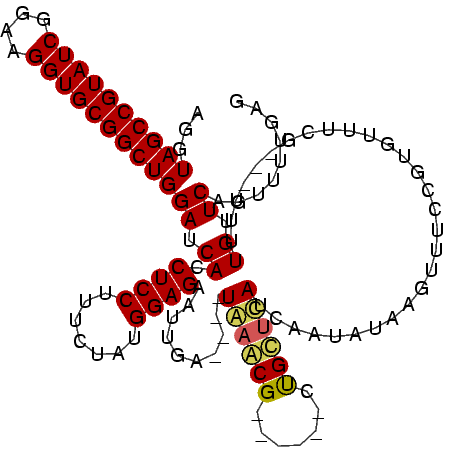
| Location | 741,570 – 741,677 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -40.13 |
| Consensus MFE | -25.56 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.64 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 107 + 5227293/1480-1600 AGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA----UAAGCG------CUGCUUAUCAAUAUAAGUUUCCGUGUUUCG---UUUUGUUCAGUUUUGAG ....(((((((((....)))))))))(((.(((((((.......)))).(((((----(((((.------..))))))))))..........))).))).---................. ( -37.30) >Bcere.0 818171 107 + 5224283/1480-1600 AGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA----UGAACG------CUGUUCAUCAAUAUAAGUUUCCGUGUUUCG---UUUUGUUCAGUUUUGAG ....(((((((((....)))))))))(((.(((((((.......)))).(((((----(((((.------..))))))))))..........))).))).---................. ( -37.20) >Bhalo.0 50621 120 + 4202352/1480-1600 AGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUACGGAGUUUUUAACUCUAGUCGGACCAUUCGCAGAUUCAUCUGCGGAGGGACGCUUCGGACUUUUGUUCAGUUUUGAG ...((((((((((....))))))))))......((((.......)))).......(((.((.((..((.((((((((....))))))))))..))))..((((....))))......))) ( -45.90) >consensus AGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA____UAAACG______CUGCUCAUCAAUAUAAGUUUCCGUGUUUCG___UUUUGUUCAGUUUUGAG ...((((((((((....))))))))))((.((.((((.......))))..........((((((.......)))))).............................)).))......... (-25.56 = -25.90 + 0.34)

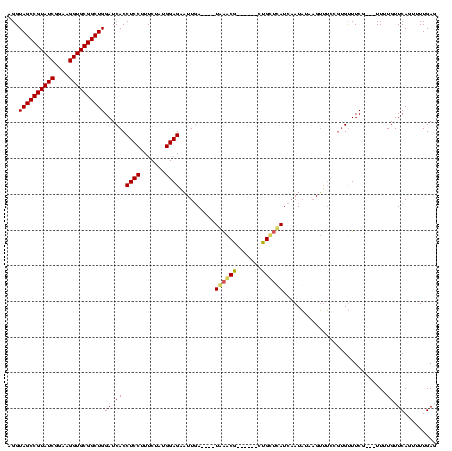
| Location | 741,570 – 741,674 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -18.93 |
| Energy contribution | -19.60 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 104 + 5227293/1497-1617 GGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA----UAAGCG------CUGCUUAUCAAUAUAAGUUUCCGUGUUUCG---UUUUGUUCAGUUUUGAGAGAACU---AUCUCUCA .....(((((((((((((((.......)))).(((((----(((((.------..))))))))))..........))).....---....)))))))).(((((((...---.))))))) ( -30.10) >Bcere.0 818171 104 + 5224283/1497-1617 GGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA----UGAACG------CUGUUCAUCAAUAUAAGUUUCCGUGUUUCG---UUUUGUUCAGUUUUGAGAGAACU---AUCUCUCA .....(((((((((((((((.......)))).(((((----(((((.------..))))))))))..........))).....---....)))))))).(((((((...---.))))))) ( -30.00) >Bhalo.0 50621 120 + 4202352/1497-1617 GGUGCGGCUGGAUCACCUCCUUUCUACGGAGUUUUUAACUCUAGUCGGACCAUUCGCAGAUUCAUCUGCGGAGGGACGCUUCGGACUUUUGUUCAGUUUUGAGAGAACUCAAAACUCUCA (((.((((((((....((((.......))))........)))))))).))).((((((((....))))))))((((......((((....)))).((((((((....)))))))))))). ( -45.80) >consensus GGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA____UAAACG______CUGCUCAUCAAUAUAAGUUUCCGUGUUUCG___UUUUGUUCAGUUUUGAGAGAACU___AUCUCUCA .....((((((((...((((.......))))..........((((((.......))))))..............................)))))))).(((((((.......))))))) (-18.93 = -19.60 + 0.67)

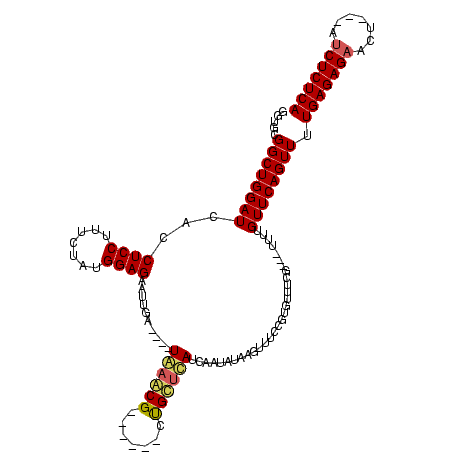
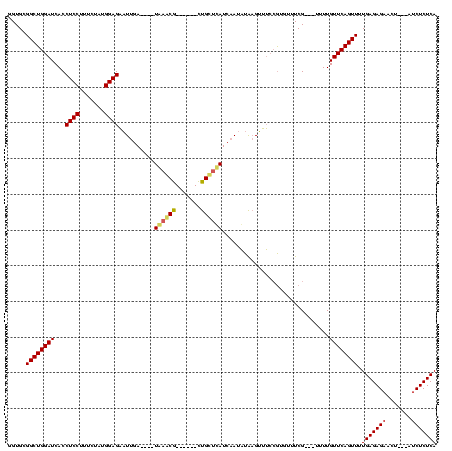
| Location | 741,570 – 741,674 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -14.44 |
| Energy contribution | -15.33 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 741570 104 + 5227293/1497-1617 UGAGAGAU---AGUUCUCUCAAAACUGAACAAAA---CGAAACACGGAAACUUAUAUUGAUAAGCAG------CGCUUA----UCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACC (((((((.---...))))))).............---........(((.......((((((((((..------.)))))----)))))(((((.......)))))...)))......... ( -27.60) >Bcere.0 818171 104 + 5224283/1497-1617 UGAGAGAU---AGUUCUCUCAAAACUGAACAAAA---CGAAACACGGAAACUUAUAUUGAUGAACAG------CGUUCA----UCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACC (((((((.---...))))))).............---........(((.......((((((((((..------.)))))----)))))(((((.......)))))...)))......... ( -27.50) >Bhalo.0 50621 120 + 4202352/1497-1617 UGAGAGUUUUGAGUUCUCUCAAAACUGAACAAAAGUCCGAAGCGUCCCUCCGCAGAUGAAUCUGCGAAUGGUCCGACUAGAGUUAAAAACUCCGUAGAAAGGAGGUGAUCCAGCCGCACC ....(((((((((....))))))))).........(((..((((..((..((((((....))))))...))..)).)).((((.....))))........)))(((......)))..... ( -33.50) >consensus UGAGAGAU___AGUUCUCUCAAAACUGAACAAAA___CGAAACACGGAAACUUAUAUUGAUAAGCAG______CGAUUA____UCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACC (((((((.......)))))))...(((.................................(((((.........)))))..........((((.......))))......)))....... (-14.44 = -15.33 + 0.89)

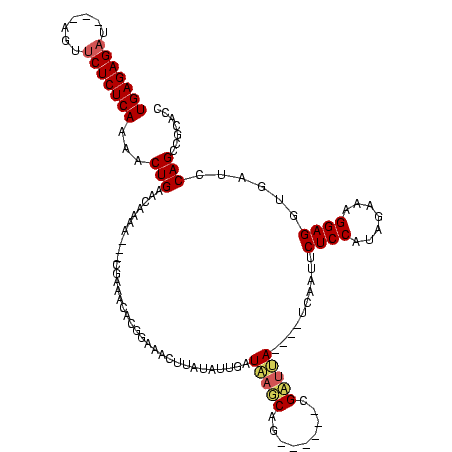

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:27:27 2006