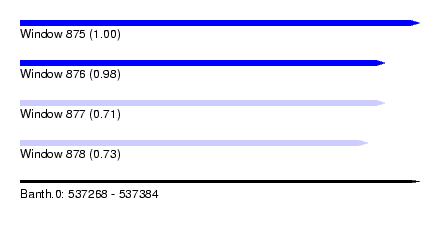
| Sequence ID | Banth.0 |
|---|---|
| Location | 537,268 – 537,384 |
| Length | 116 |
| Max. P | 0.999265 |

| Location | 537,268 – 537,384 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.62 |
| Mean single sequence MFE | -39.78 |
| Consensus MFE | -37.91 |
| Energy contribution | -36.97 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 537268 116 + 5227293/0-120 CAAUCGUGCUCCUUCGGCCAACUCGGACAGCUCUCC----AAUGGCUCCGCAGGUAGGAAUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUACCGCUGAGCUACUGCGGAAUA ................((((....(((......)))----..))))((((((((((((.......))))).....((((((.....))))))(((((.......))))).)))))))... ( -39.30) >Bcere.0 609771 116 + 5224283/0-120 CAAUCGUGCUCCUUCGGCCAACUCGGACAGCUCUCC----AAUGGCUCCGCAGGUAGGAAUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUACCGCUGAGCUACUGCGGAAUA ................((((....(((......)))----..))))((((((((((((.......))))).....((((((.....))))))(((((.......))))).)))))))... ( -39.30) >Bhalo.0 1023793 115 + 4202352/0-120 CAAUCGUGCUCCUUCGGCCAACUCGGACAGCUCUCC-----GUGGCUCCACAGGUAGGACUCGAACCUACGACCGAUCGGUUAACAGCCGAUUGCUCUACCACUGAGCUACUGUGGAACA .....((.........((((...((((......)))-----)))))((((((((((((.......)))))....(((((((.....)))))))((((.......))))..))))))))). ( -40.20) >Bclau.0 1446200 117 + 4303871/0-120 CAAUCGUGCUCCUUCGGCCAACUCGGACAGCUCUCC---UAAUGGCUCCACAGGCAGGAUUCGAACCUGCGACCGAUCGGUUAACAGCCGAUAGCUCUACCACUGAGCUACUGUGGAACG ....(((.........((((....(((......)))---...))))((((((((((((.......))))).....((((((.....))))))(((((.......))))).)))))))))) ( -40.70) >Bsubt.0 950912 118 + 4214630/0-120 CAAUCGUGCUCCUUCGACC-ACUCGGACAGCUCUCC-AAUAAUGGCUCCGCAGGUAGGAUUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUACCACUGAGCUACUGCGGAAUG .....(.(((..(((((..-..))))).))).).((-(....))).((((((((((((.......))))).....((((((.....))))))(((((.......))))).)))))))... ( -38.60) >Blich.0 925432 119 + 4222334/0-120 CAAUCGUGCUCCUUCGACC-ACUCGGACAGCUCUCCAAUAAAUGGCUCCGCAGGCAGGAUUCGAACCUGCGACCGAUCGGUUAACAGCCGAUAGCUCUACCACUGAGCUACUGCGGAAUA .....(.(((..(((((..-..))))).))).).(((.....))).((((((((((((.......))))).....((((((.....))))))(((((.......))))).)))))))... ( -40.60) >consensus CAAUCGUGCUCCUUCGGCCAACUCGGACAGCUCUCC____AAUGGCUCCGCAGGUAGGAUUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUACCACUGAGCUACUGCGGAAUA .....(.(((..(((((.....))))).))).).............((((((((((((.......))))).....((((((.....))))))(((((.......))))).)))))))... (-37.91 = -36.97 + -0.94)

| Location | 537,268 – 537,374 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.31 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -37.56 |
| Energy contribution | -35.78 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 537268 106 + 5227293/80-200 ----GGAGAGCUGUCCGAGUUGGCCGAAGGAGCACGAUUGGAAAUCGUGUAUACGUCAC--AAGCGUAUCAAGGGUUCGAAUCCCUUGCUCUCCGCCAUAGG-----AUUUAUAAUA--- ----(((((((..(((.((((((((....).)).))))))))......(.((((((...--..)))))))(((((.......))))))))))))........-----..........--- ( -33.80) >Bcere.0 609771 106 + 5224283/80-200 ----GGAGAGCUGUCCGAGUUGGCCGAAGGAGCACGAUUGGAAAUCGUGUAUACGUCAC--AAGCGUAUCAAGGGUUCGAAUCCCUUGCUCUCCGCCAUAGG-----AUUUAUAAUA--- ----(((((((..(((.((((((((....).)).))))))))......(.((((((...--..)))))))(((((.......))))))))))))........-----..........--- ( -33.80) >Bhalo.0 1023793 104 + 4202352/80-200 ----GGAGAGCUGUCCGAGUUGGCCGAAGGAGCACGAUUGGAAAUCGUGUAGGCGGUGACAAACCGUCUCGAGGGUUCGAAUCCCUCGCUCUCCGCCAG--------AUAAAUUAC---- ----(((((((..(((.((((((((....).)).))))))))......(.(((((((.....))))))))(((((.......)))))))))))).....--------.........---- ( -42.80) >Bclau.0 1446200 105 + 4303871/80-200 A---GGAGAGCUGUCCGAGUUGGCCGAAGGAGCACGAUUGGAAAUCGUGUAGGCGGUGUAAAACCGUCUCGAGGGUUCGAAUCCCUCGCUCUCCGCCAUC-------AUACAUAA----- .---(((((((..(((.((((((((....).)).))))))))......(.(((((((.....))))))))(((((.......))))))))))))......-------........----- ( -43.00) >Bsubt.0 950912 117 + 4214630/80-200 AUU-GGAGAGCUGUCCGAGU-GGUCGAAGGAGCACGAUUGGAAAUCGUGUAGGCGGUCA-ACUCCGUCUCAAGGGUUCGAAUCCCUUGCUCUCCGCCACUGAUACUUAUACAUAAUUACA ...-(((((((.......((-(.((....)).)))((((.(((..(.((.((((((...-...)))))))).)..))).))))....))))))).......................... ( -39.50) >Blich.0 925432 108 + 4222334/80-200 UAUUGGAGAGCUGUCCGAGU-GGUCGAAGGAGCACGAUUGGAAAUCGUGUAGGCGGCCA-ACGCCGUCUCAAGGGUUCGAAUCCCUUGCUCUCCGCCACU-------UUAAAUACAU--- ....(((((((.......((-(.((....)).)))((((.(((..(.((.(((((((..-..))))))))).)..))).))))....)))))))......-------..........--- ( -42.10) >consensus ____GGAGAGCUGUCCGAGUUGGCCGAAGGAGCACGAUUGGAAAUCGUGUAGGCGGCAA_AAACCGUCUCAAGGGUUCGAAUCCCUUGCUCUCCGCCAUA_______AUAAAUAAU____ ....(((((((..(((((....(((....).))....)))))......(.((((((.......)))))))(((((.......)))))))))))).......................... (-37.56 = -35.78 + -1.77)

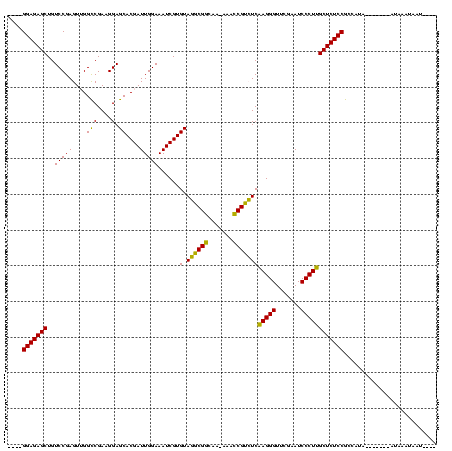
| Location | 537,268 – 537,374 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.31 |
| Mean single sequence MFE | -33.25 |
| Consensus MFE | -30.07 |
| Energy contribution | -29.18 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 537268 106 + 5227293/80-200 ---UAUUAUAAAU-----CCUAUGGCGGAGAGCAAGGGAUUCGAACCCUUGAUACGCUU--GUGACGUAUACACGAUUUCCAAUCGUGCUCCUUCGGCCAACUCGGACAGCUCUCC---- ---.........(-----((..((((.(((((((((((.......))))))((((((..--..).))))).((((((.....)))))))))..)).))))....))).........---- ( -29.30) >Bcere.0 609771 106 + 5224283/80-200 ---UAUUAUAAAU-----CCUAUGGCGGAGAGCAAGGGAUUCGAACCCUUGAUACGCUU--GUGACGUAUACACGAUUUCCAAUCGUGCUCCUUCGGCCAACUCGGACAGCUCUCC---- ---.........(-----((..((((.(((((((((((.......))))))((((((..--..).))))).((((((.....)))))))))..)).))))....))).........---- ( -29.30) >Bhalo.0 1023793 104 + 4202352/80-200 ----GUAAUUUAU--------CUGGCGGAGAGCGAGGGAUUCGAACCCUCGAGACGGUUUGUCACCGCCUACACGAUUUCCAAUCGUGCUCCUUCGGCCAACUCGGACAGCUCUCC---- ----.........--------.....((((((((((((.......)))))(((..(((.....)))(((..((((((.....)))))).......)))...))).....)))))))---- ( -34.90) >Bclau.0 1446200 105 + 4303871/80-200 -----UUAUGUAU-------GAUGGCGGAGAGCGAGGGAUUCGAACCCUCGAGACGGUUUUACACCGCCUACACGAUUUCCAAUCGUGCUCCUUCGGCCAACUCGGACAGCUCUCC---U -----........-------......((((((((((((.......)))))(((..(((.....)))(((..((((((.....)))))).......)))...))).....)))))))---. ( -36.10) >Bsubt.0 950912 117 + 4214630/80-200 UGUAAUUAUGUAUAAGUAUCAGUGGCGGAGAGCAAGGGAUUCGAACCCUUGAGACGGAGU-UGACCGCCUACACGAUUUCCAAUCGUGCUCCUUCGACC-ACUCGGACAGCUCUCC-AAU ....................((((((((((..((((((.......))))))((.(((...-...))).)).((((((.....))))))...))))).))-))).(((......)))-... ( -33.10) >Blich.0 925432 108 + 4222334/80-200 ---AUGUAUUUAA-------AGUGGCGGAGAGCAAGGGAUUCGAACCCUUGAGACGGCGU-UGGCCGCCUACACGAUUUCCAAUCGUGCUCCUUCGACC-ACUCGGACAGCUCUCCAAUA ---..........-------((((((((((..((((((.......))))))((.((((..-..)))).)).((((((.....))))))...))))).))-))).(((......))).... ( -36.80) >consensus ____AUUAUAUAU_______AAUGGCGGAGAGCAAGGGAUUCGAACCCUUGAGACGGUUU_UCACCGCCUACACGAUUUCCAAUCGUGCUCCUUCGGCCAACUCGGACAGCUCUCC____ ..........................((((((((((((.......))))).((.((((.....)))).)).((((((.....))))))....(((((.....)))))..))))))).... (-30.07 = -29.18 + -0.88)

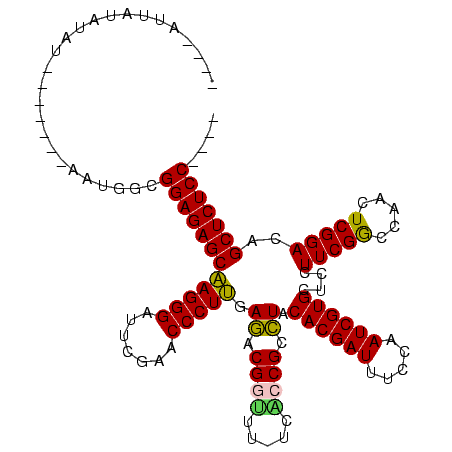
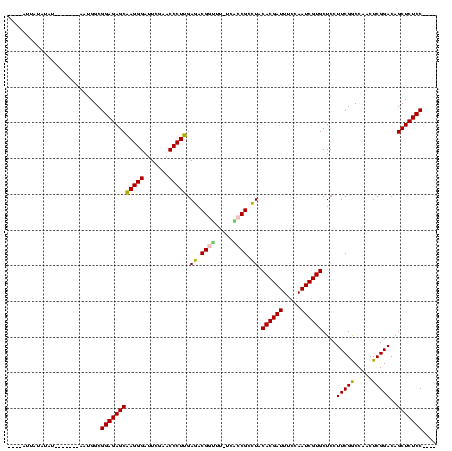
| Location | 537,268 – 537,369 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -30.34 |
| Consensus MFE | -24.32 |
| Energy contribution | -22.72 |
| Covariance contribution | -1.61 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 537268 101 + 5227293/120-240 GAAAUCGUGUAUACGUCAC--AAGCGUAUCAAGGGUUCGAAUCCCUUGCUCUCCGCCAUAGG-----AUUUAUAAUA------------UUUGGCCCGUUGGUCAAGUGGUUAAGACACC ......((((((((((...--..))))))((((((.......))))))...(((......))-----)((((...((------------(((((((....)))))))))..)))))))). ( -27.20) >Bcere.0 609771 101 + 5224283/120-240 GAAAUCGUGUAUACGUCAC--AAGCGUAUCAAGGGUUCGAAUCCCUUGCUCUCCGCCAUAGG-----AUUUAUAAUA------------UUUGGCCCGUUGGUCAAGUGGUUAAGACACC ......((((((((((...--..))))))((((((.......))))))...(((......))-----)((((...((------------(((((((....)))))))))..)))))))). ( -27.20) >Bhalo.0 1023793 99 + 4202352/120-240 GAAAUCGUGUAGGCGGUGACAAACCGUCUCGAGGGUUCGAAUCCCUCGCUCUCCGCCAG--------AUAAAUUAC-------------CGUGGCCCGUUGGUCAAGUGGUUAAGACACC ......((((.(((((.(((.....))).((((((.......))))))....)))))..--------...((((((-------------..(((((....))))).))))))...)))). ( -34.00) >Bclau.0 1446200 98 + 4303871/120-240 GAAAUCGUGUAGGCGGUGUAAAACCGUCUCGAGGGUUCGAAUCCCUCGCUCUCCGCCAUC-------AUACAUAA---------------AAGGCCCGUUGGUCAAGCGGUUAAGACACC ......(((((((((((.....)))))))((((((.......))))))......(((...-------........---------------..)))(((((.....))))).....)))). ( -30.02) >Bsubt.0 950912 119 + 4214630/120-240 GAAAUCGUGUAGGCGGUCA-ACUCCGUCUCAAGGGUUCGAAUCCCUUGCUCUCCGCCACUGAUACUUAUACAUAAUUACAAUUGAAAGUCUGGGCCCGUUGGUCAAGCGGUUAAGACACC (.(((..(((((((((...-...))))).((((((.......))))))....................))))..))).)........((((..(((.(((.....))))))..))))... ( -30.50) >Blich.0 925432 99 + 4222334/120-240 GAAAUCGUGUAGGCGGCCA-ACGCCGUCUCAAGGGUUCGAAUCCCUUGCUCUCCGCCACU-------UUAAAUACAU-------------CUGGCCCGUUGGUCAAGUGGUUAAGACACC ......(((((((((((..-..)))))))((((((.......))))))..((..((((((-------..........-------------.(((((....)))))))))))..)))))). ( -33.10) >consensus GAAAUCGUGUAGGCGGCAA_AAACCGUCUCAAGGGUUCGAAUCCCUUGCUCUCCGCCAUA_______AUAAAUAAU______________UUGGCCCGUUGGUCAAGUGGUUAAGACACC ......((((((((((.......))))))((((((.......))))))((..((((............(....)..................((((....))))..))))...)))))). (-24.32 = -22.72 + -1.61)

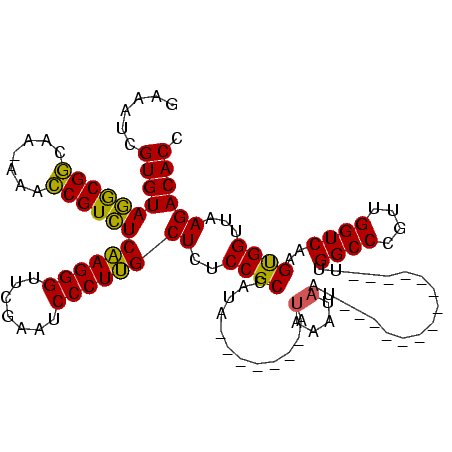
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:27:04 2006