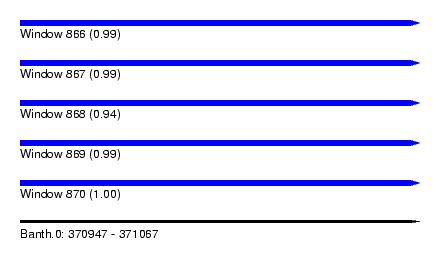
| Sequence ID | Banth.0 |
|---|---|
| Location | 370,947 – 371,067 |
| Length | 120 |
| Max. P | 0.997862 |

| Location | 370,947 – 371,067 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 59.52 |
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -10.80 |
| Energy contribution | -11.28 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.40 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 370947 120 + 5227293/0-120 UAUGUUUUGAAAUAUAAAAAAACUCACCUUUCAUGAUCAAGAAAGAUGAGUUUUGUACACAAUAGUUACGUUUUAUCUUUCAAAAUGCAAUGCAUUUUGCAGGAAUUAGCACCUAUUCAG ...(((((((((.(((((((((((((.(((((........))))).))))))))(((.((....)))))..))))).)))))))))((((......))))(((........)))...... ( -27.90) >Bcere.0 470944 119 + 5224283/0-120 UAUAUUUUGAAAUAUAAAAAA-CUCACCUUUCAUAAUCAAGAAAGAUGAGUUUUGCACACAAUAGUUACGUUUUAUCUUUCAAAAUGCAAUGCAUUUUGCAGGAAUUAGCACCUAUUCAG .......((((......((((-((((.(((((........))))).))))))))...............(((...((((.((((((((...)))))))).))))...))).....)))). ( -27.10) >Bsubt.0 2585051 109 - 4214630/0-120 UACCUCUUAAAAUAAAAAACUUUCAAUCCGGCG--GA-AAGAAAGACAGGA-------ACAGCAUGCAGCCCUUAUCUUUCAAAAUGGAAUUCAUUUUGCUGGAAGUAGCACCU-UACAA ..........................((((((.--..-..((((((.(((.-------...((.....)))))..))))))((((((.....)))))))))))).((((....)-))).. ( -20.60) >Blich.0 2487439 110 - 4222334/0-120 UGCCCCUAAAAAAUUAAAAUCUUUCUUCCAAUUUUGA-AAGAAAGACGAGG-------AACUCCGCUAUUCCUUAUCUCUCAAAAUGAGGG-CAUUUUGCUGGAAGUAGCACCU-UACGC (((((..............((((((..........))-)))).(((..(((-------((........)))))..)))..........)))-))...(((((....)))))...-..... ( -24.80) >Bhalo.0 910027 100 + 4202352/0-120 AAGGUGUUAAAACAAGAAAAAACACCUCUUCGAU-----AAGAAGACGUGC-------GAUCCGUUCGCUUCUUAUCUUUCAGAACAUCAU--GUUCUGCUGGAAUUGGCACCU------ .(((((((((...(((((.....((.(((((...-----..))))).))((-------((.....))))))))).(((..((((((.....--))))))..))).)))))))))------ ( -33.80) >consensus UACCUCUUAAAAUAUAAAAAAACUCAUCUUUCAU_AA_AAGAAAGACGAGU_______ACAACAGUUACGUCUUAUCUUUCAAAAUGCAAU_CAUUUUGCUGGAAUUAGCACCU_UACAG ......................(((.((((((........)))))).))).........................(((..(((((((.....)))))))..)))................ (-10.80 = -11.28 + 0.48)


| Location | 370,947 – 371,067 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 59.52 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -13.00 |
| Energy contribution | -13.68 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 370947 120 + 5227293/0-120 CUGAAUAGGUGCUAAUUCCUGCAAAAUGCAUUGCAUUUUGAAAGAUAAAACGUAACUAUUGUGUACAAAACUCAUCUUUCUUGAUCAUGAAAGGUGAGUUUUUUUAUAUUUCAAAACAUA ......(((........)))((((......)))).((((((((.(((((..(((((....)).)))((((((((((((((........))))))))))))))))))).)))))))).... ( -30.60) >Bcere.0 470944 119 + 5224283/0-120 CUGAAUAGGUGCUAAUUCCUGCAAAAUGCAUUGCAUUUUGAAAGAUAAAACGUAACUAUUGUGUGCAAAACUCAUCUUUCUUGAUUAUGAAAGGUGAG-UUUUUUAUAUUUCAAAAUAUA ...(((((.(((....((...((((((((...))))))))...))......))).)))))(((((.((((((((((((((........))))))))))-)))).)))))........... ( -32.10) >Bsubt.0 2585051 109 - 4214630/0-120 UUGUA-AGGUGCUACUUCCAGCAAAAUGAAUUCCAUUUUGAAAGAUAAGGGCUGCAUGCUGU-------UCCUGUCUUUCUU-UC--CGCCGGAUUGAAAGUUUUUUAUUUUAAGAGGUA ..(((-(((.(((...(((.((((((((.....))))))((((((((.((((........))-------)).))))))))..-..--.)).))).....))).))))))........... ( -23.90) >Blich.0 2487439 110 - 4222334/0-120 GCGUA-AGGUGCUACUUCCAGCAAAAUG-CCCUCAUUUUGAGAGAUAAGGAAUAGCGGAGUU-------CCUCGUCUUUCUU-UCAAAAUUGGAAGAAAGAUUUUAAUUUUUUAGGGGCA ((..(-((......)))...))....((-((((.....((((((((.((((((......)))-------))).(((((((((-(((....))))))))))))....)))))))))))))) ( -33.00) >Bhalo.0 910027 100 + 4202352/0-120 ------AGGUGCCAAUUCCAGCAGAAC--AUGAUGUUCUGAAAGAUAAGAAGCGAACGGAUC-------GCACGUCUUCUU-----AUCGAAGAGGUGUUUUUUCUUGUUUUAACACCUU ------(((((..........((((((--.....)))))).((((((((((((((.....))-------))((.(((((..-----...))))).)).....))))))))))..))))). ( -32.80) >consensus CUGAA_AGGUGCUAAUUCCAGCAAAAUG_AUUGCAUUUUGAAAGAUAAGAAGUAACUGUUGU_______ACUCGUCUUUCUU_AC_AUGAAAGAUGAGAUUUUUUAUAUUUUAAAACAUA .......((........))..(((((((.....))))))).............................(((((((((((........)))))))))))..................... (-13.00 = -13.68 + 0.68)

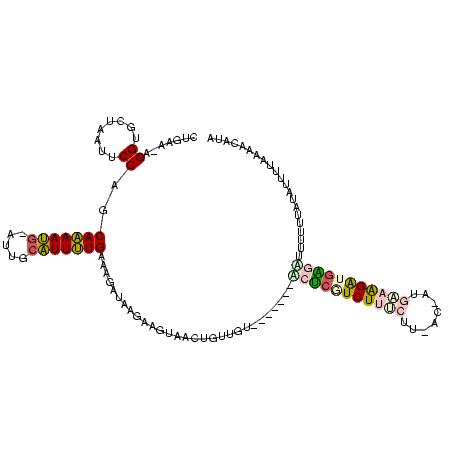
| Location | 370,947 – 371,067 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.52 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -18.19 |
| Energy contribution | -17.71 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 370947 120 + 5227293/40-160 GAAAGAUGAGUUUUGUACACAAUAGUUACGUUUUAUCUUUCAAAAUGCAAUGCAUUUUGCAGGAAUUAGCACCUAUUCAGUUUGUAUAAACUGAGGUUGCUGGGCUUCAAAGGGCCGGUC ((((((((((...((((.((....)))))).))))))))))....(((((......))))).....(((((((...(((((((....))))))))).)))))((((......)))).... ( -34.60) >Bcere.0 470944 120 + 5224283/40-160 GAAAGAUGAGUUUUGCACACAAUAGUUACGUUUUAUCUUUCAAAAUGCAAUGCAUUUUGCAGGAAUUAGCACCUAUUCAGUUUGUAUAAACUGAGGUUGCUGGGCUUCAAAGGGCCGGUC ((((((((((....((........)).....))))))))))....(((((......))))).....(((((((...(((((((....))))))))).)))))((((......)))).... ( -32.70) >Bsubt.0 2585051 106 - 4214630/40-160 GAAAGACAGGA-------ACAGCAUGCAGCCCUUAUCUUUCAAAAUGGAAUUCAUUUUGCUGGAAGUAGCACCU-UACAAACUAU------GCAGGUUGCCGGGCUUCAUAGGUCCAGUC ....(((.(((-------.(...(((.(((((...(((..(((((((.....)))))))..)))....((((((-..((.....)------).))).))).))))).)))..)))).))) ( -28.40) >Blich.0 2487439 107 - 4222334/40-160 GAAAGACGAGG-------AACUCCGCUAUUCCUUAUCUCUCAAAAUGAGGG-CAUUUUGCUGGAAGUAGCACCU-UACGCUUCACA----CGCAGGUUGCCGGGUUUCAUCGGACCAGUC ((.(((..(((-------((........)))))..))).))........((-((.((((((((((((.......-...))))).))----.))))).)))).((((......)))).... ( -31.10) >Bhalo.0 910027 105 + 4202352/40-160 AGAAGACGUGC-------GAUCCGUUCGCUUCUUAUCUUUCAGAACAUCAU--GUUCUGCUGGAAUUGGCACCU------UCUUUAAAAAAAGAGGUUGCCGGGCUUCACAGGGCCAGUC (((((.((.((-------(...))).)))))))..(((..((((((.....--))))))..)))..(((((.((------(((((....))))))).)))))((((......)))).... ( -36.30) >consensus GAAAGACGAGU_______ACAACAGUUACGUCUUAUCUUUCAAAAUGCAAU_CAUUUUGCUGGAAUUAGCACCU_UACAGUUUAUA_AAACGGAGGUUGCCGGGCUUCAAAGGGCCAGUC ....(((............................(((..(((((((.....)))))))..)))..((((((((...................))).)))))((((......)))).))) (-18.19 = -17.71 + -0.48)

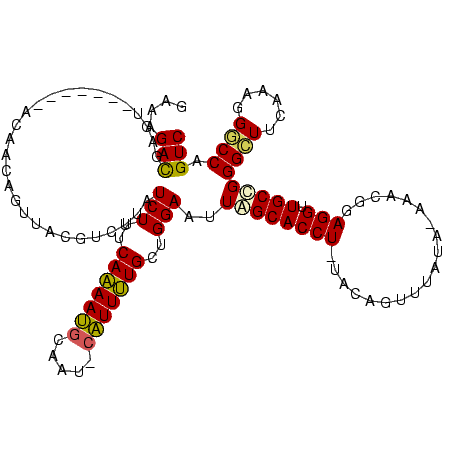
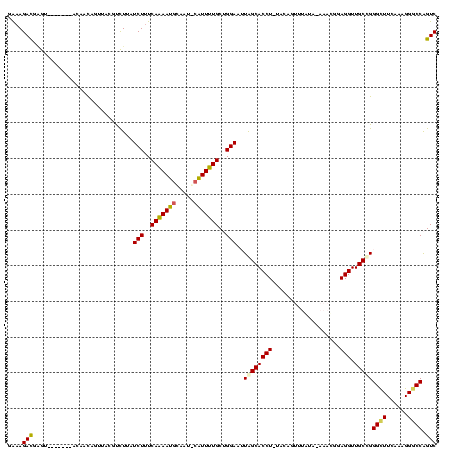
| Location | 370,947 – 371,067 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.98 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -21.77 |
| Energy contribution | -20.49 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 370947 120 + 5227293/68-188 CGUUUUAUCUUUCAAAAUGCAAUGCAUUUUGCAGGAAUUAGCACCUAUUCAGUUUGUAUAAACUGAGGUUGCUGGGCUUCAAAGGGCCGGUCCCUCUGCCUCUCUUGAUAAAGAAUUAUU .......(((((((((((((...))))))))(((((..(((((((...(((((((....))))))))).)))))(((.....((((.....))))..)))..)))))..)))))...... ( -32.70) >Bcere.0 470944 120 + 5224283/68-188 CGUUUUAUCUUUCAAAAUGCAAUGCAUUUUGCAGGAAUUAGCACCUAUUCAGUUUGUAUAAACUGAGGUUGCUGGGCUUCAAAGGGCCGGUCCCUCUGCCUCUCUUGAUAAAGGUAUAUU .....(((((((((((((((...))))))))(((((..(((((((...(((((((....))))))))).)))))(((.....((((.....))))..)))..)))))..))))))).... ( -34.20) >Bsubt.0 2585051 113 - 4214630/68-188 GCCCUUAUCUUUCAAAAUGGAAUUCAUUUUGCUGGAAGUAGCACCU-UACAAACUAU------GCAGGUUGCCGGGCUUCAUAGGUCCAGUCCCUCAGCCGCUCUUGAUAAGUUUUUAGA ...((((((...(((((((.....)))))))....(((.(((....-..((.....)------)..(((((..(((((..........)))))..)))))))))))))))))........ ( -24.20) >Blich.0 2487439 114 - 4222334/68-188 UUCCUUAUCUCUCAAAAUGAGGG-CAUUUUGCUGGAAGUAGCACCU-UACGCUUCACA----CGCAGGUUGCCGGGUUUCAUCGGACCAGUCCCUCCACCGCUCUUGAUAAGUUUGCUUU ...((((((.(((.....)))((-((.((((((((((((.......-...))))).))----.))))).))))((((......(((....))).......))))..))))))........ ( -29.22) >Bhalo.0 910027 112 + 4202352/68-188 CUUCUUAUCUUUCAGAACAUCAU--GUUCUGCUGGAAUUGGCACCU------UCUUUAAAAAAAGAGGUUGCCGGGCUUCACAGGGCCAGUCCCUCCACCACUCUGGAUAAGAAAAUAUG .(((((((((..((((((.....--)))))).((((..(((((.((------(((((....))))))).)))))((((......))))......)))).......)))))))))...... ( -38.10) >consensus CGUCUUAUCUUUCAAAAUGCAAU_CAUUUUGCUGGAAUUAGCACCU_UACAGUUUAUA_AAACGGAGGUUGCCGGGCUUCAAAGGGCCAGUCCCUCCGCCGCUCUUGAUAAGGAUAUAUU ...((((((...(((((((.....))))))).(((...((((((((...................))).)))))((((......))))..........))).....))))))........ (-21.77 = -20.49 + -1.28)


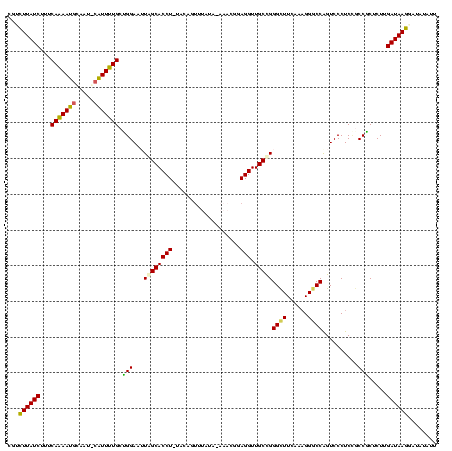
| Location | 370,947 – 371,067 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.98 |
| Mean single sequence MFE | -33.72 |
| Consensus MFE | -23.27 |
| Energy contribution | -21.83 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 370947 120 + 5227293/68-188 AAUAAUUCUUUAUCAAGAGAGGCAGAGGGACCGGCCCUUUGAAGCCCAGCAACCUCAGUUUAUACAAACUGAAUAGGUGCUAAUUCCUGCAAAAUGCAUUGCAUUUUGAAAGAUAAAACG ........((((((.....((((((((((.....)))))))......((((.(((((((((....)))))))...))))))....))).((((((((...))))))))...))))))... ( -34.30) >Bcere.0 470944 120 + 5224283/68-188 AAUAUACCUUUAUCAAGAGAGGCAGAGGGACCGGCCCUUUGAAGCCCAGCAACCUCAGUUUAUACAAACUGAAUAGGUGCUAAUUCCUGCAAAAUGCAUUGCAUUUUGAAAGAUAAAACG ........((((((.....((((((((((.....)))))))......((((.(((((((((....)))))))...))))))....))).((((((((...))))))))...))))))... ( -34.30) >Bsubt.0 2585051 113 - 4214630/68-188 UCUAAAAACUUAUCAAGAGCGGCUGAGGGACUGGACCUAUGAAGCCCGGCAACCUGC------AUAGUUUGUA-AGGUGCUACUUCCAGCAAAAUGAAUUCCAUUUUGAAAGAUAAGGGC ........((((((((((((((((.(((.......)))....)))).....((((((------(.....))).-))))))).)))....(((((((.....)))))))...))))))... ( -30.90) >Blich.0 2487439 114 - 4222334/68-188 AAAGCAAACUUAUCAAGAGCGGUGGAGGGACUGGUCCGAUGAAACCCGGCAACCUGCG----UGUGAAGCGUA-AGGUGCUACUUCCAGCAAAAUG-CCCUCAUUUUGAGAGAUAAGGAA ........((((((......(.((((((....(((........))).((((.((((((----(.....)))).-))))))).)))))).)......-..(((.....))).))))))... ( -32.30) >Bhalo.0 910027 112 + 4202352/68-188 CAUAUUUUCUUAUCCAGAGUGGUGGAGGGACUGGCCCUGUGAAGCCCGGCAACCUCUUUUUUUAAAGA------AGGUGCCAAUUCCAGCAGAAC--AUGAUGUUCUGAAAGAUAAGAAG ......((((((((........(((((((.((..........)))))((((.(((((((....)))).------)))))))...)))).((((((--.....))))))...)))))))). ( -36.80) >consensus AAUAAAAACUUAUCAAGAGAGGCAGAGGGACUGGCCCUAUGAAGCCCGGCAACCUCAGUUU_UACAAACUGAA_AGGUGCUAAUUCCAGCAAAAUG_AUUGCAUUUUGAAAGAUAAGAAG ........((((((....(.(((..((((.....)))).....))))((((.(((...................)))))))........(((((((.....)))))))...))))))... (-23.27 = -21.83 + -1.44)

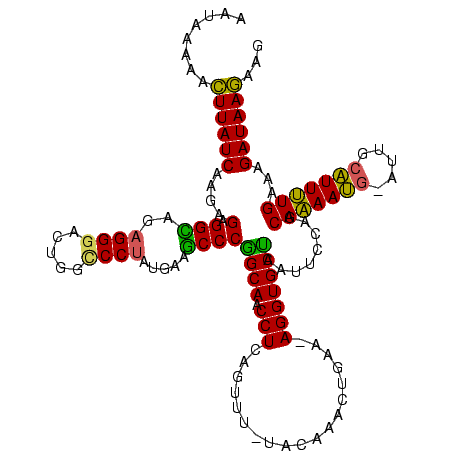
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:26:57 2006