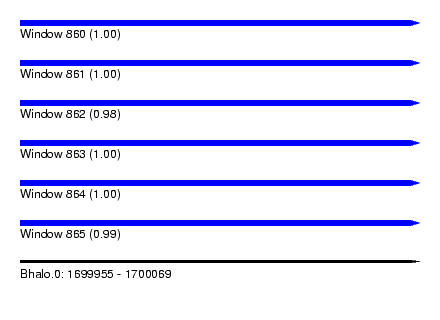
| Sequence ID | Bhalo.0 |
|---|---|
| Location | 1,699,955 – 1,700,069 |
| Length | 114 |
| Max. P | 0.999966 |

| Location | 1,699,955 – 1,700,069 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.76 |
| Mean single sequence MFE | -38.55 |
| Consensus MFE | -31.89 |
| Energy contribution | -31.82 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.83 |
| SVM decision value | 4.98 |
| SVM RNA-class probability | 0.999966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 1699955 114 + 4202352/0-120 AUUCUUAUCCAGAGAGGUGGAGGGAACGGCCCGAAGAAACCUCAGCAACCAGCCACGAUCCUGUGGUCAGGUGCUAAUUCCUGCAAGCAUUAUUUG--CUUGAGAGAUAAGAGGA----A .(((((((((((.(((((...(((.....)))......)))))((((.((.((((((....))))))..)))))).....)))((((((.....))--))))...))))))))..----. ( -42.00) >Bclau.0 1983210 103 + 4303871/0-120 GUUCUUAUCAAGAGAAGUGGAGGGAAUGGCCCAAUGAAGCUUCAGCAACCAGCCAA----CCAUGGUCAGGUGCUAAAUCCAGCAG------UUUA--UCUGCAAGAUAAGAG-A----A .((((((((........(((((((.....)))...........((((.((.((((.----...))))..))))))...))))((((------....--.))))..))))))))-.----. ( -32.00) >Banth.0 320603 115 + 5227293/0-120 AUUCUUAUCCAGAGAGGUGGAGGGACUGGCCCUACGAUACCUCAGCAACGGGUUUU-----UUUAAUACCGUGCUAACUCCAGCAAGCCAUAUAAAGGCUUGGAAGAUGAGAAGAUGUGA .((((((((....((((((.((((.....))))....))))))((((.(((.....-----.......)))))))........((((((.......))))))...))))))))....... ( -39.40) >Bcere.0 361552 115 + 5224283/0-120 AUUCUUAUCCAGAGAGGUGGAGGGACUGGCCCUACGAUACCUCAGCAACGGGUUUU-----UUUAAUACCGUGCUAACUCCAGCAAGCCUAUGAAAGGCUUGGAAGAUGAGAAGAUGUGA .((((((((....((((((.((((.....))))....))))))((((.(((.....-----.......)))))))........(((((((.....)))))))...))))))))....... ( -40.80) >consensus AUUCUUAUCCAGAGAGGUGGAGGGAAUGGCCCUACGAAACCUCAGCAACCAGCCAU_____UUUAAUAACGUGCUAACUCCAGCAAGCCUUAUAAA__CUUGGAAGAUAAGAAGA____A .((((((((....((((((.((((.....))))....))))))((((.(((((((........)))).)))))))........((((((.......))))))...))))))))....... (-31.89 = -31.82 + -0.06)

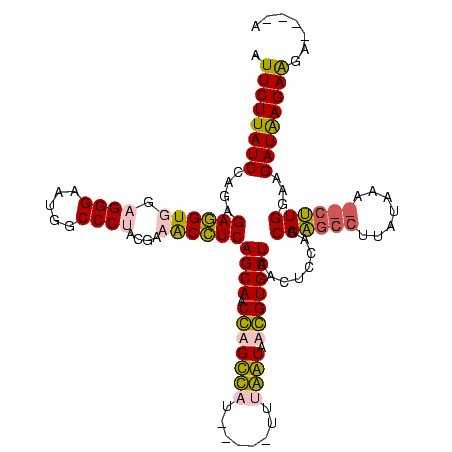
| Location | 1,699,955 – 1,700,069 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.76 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -24.52 |
| Energy contribution | -25.52 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.74 |
| SVM decision value | 3.87 |
| SVM RNA-class probability | 0.999671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 1699955 114 + 4202352/0-120 U----UCCUCUUAUCUCUCAAG--CAAAUAAUGCUUGCAGGAAUUAGCACCUGACCACAGGAUCGUGGCUGGUUGCUGAGGUUUCUUCGGGCCGUUCCCUCCACCUCUCUGGAUAAGAAU .----...(((((((...((((--((.....))))))((((....((((((.(.((((......))))).)).))))(((((......(((.....)))...)))))))))))))))).. ( -36.70) >Bclau.0 1983210 103 + 4303871/0-120 U----U-CUCUUAUCUUGCAGA--UAAA------CUGCUGGAUUUAGCACCUGACCAUGG----UUGGCUGGUUGCUGAAGCUUCAUUGGGCCAUUCCCUCCACUUCUCUUGAUAAGAAC .----.-.(((((((..((((.--....------))))(((((((((((((.(.(((...----.)))).)).)))))))........(((.....)))))))........))))))).. ( -30.90) >Banth.0 320603 115 + 5227293/0-120 UCACAUCUUCUCAUCUUCCAAGCCUUUAUAUGGCUUGCUGGAGUUAGCACGGUAUUAAA-----AAAACCCGUUGCUGAGGUAUCGUAGGGCCAGUCCCUCCACCUCUCUGGAUAAGAAU .....((((.((.(((..((((((.......))))))..)))...(((((((.......-----.....))).))))(((((...(.((((.....)))).))))))....)).)))).. ( -31.80) >Bcere.0 361552 115 + 5224283/0-120 UCACAUCUUCUCAUCUUCCAAGCCUUUCAUAGGCUUGCUGGAGUUAGCACGGUAUUAAA-----AAAACCCGUUGCUGAGGUAUCGUAGGGCCAGUCCCUCCACCUCUCUGGAUAAGAAU .....((((.((.(((..(((((((.....)))))))..)))...(((((((.......-----.....))).))))(((((...(.((((.....)))).))))))....)).)))).. ( -33.00) >consensus U____UCCUCUCAUCUUCCAAG__UAAAUAAGGCUUGCUGGAGUUAGCACCGGACCAAA_____AAAACCCGUUGCUGAGGUAUCGUAGGGCCAGUCCCUCCACCUCUCUGGAUAAGAAU ........(((((((...((((((.......))))))..(((.((((((((((.(((........))))))).)))))).........(((.....)))))).........))))))).. (-24.52 = -25.52 + 1.00)

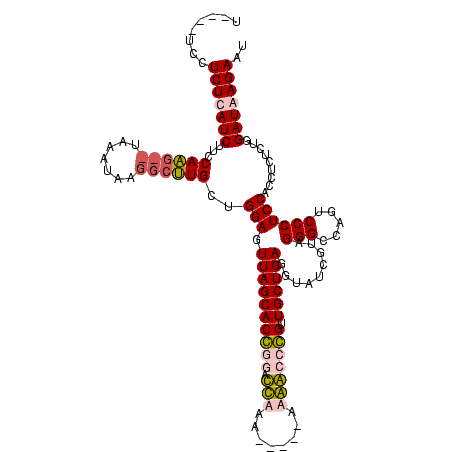
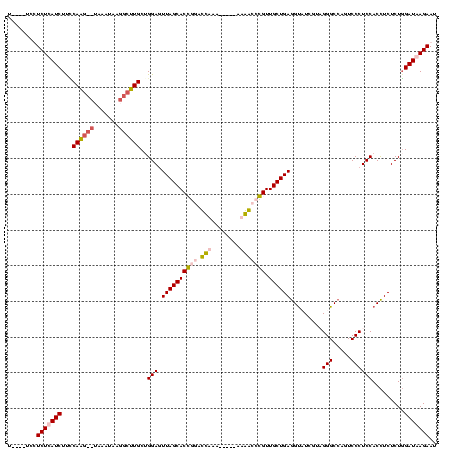
| Location | 1,699,955 – 1,700,069 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 58.20 |
| Mean single sequence MFE | -36.17 |
| Consensus MFE | -14.31 |
| Energy contribution | -17.62 |
| Covariance contribution | 3.31 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.88 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 1699955 114 + 4202352/40-160 CUCAGCAACCAGCCACGAUCCUGUGGUCAGGUGCUAAUUCCUGCAAGCAUUAUUUG--CUUGAGAGAUAAGAGGA----AGCGAGUGAGAUCCAACACCUACUUCUUCUUAAUCUUACAU ...((((.((.((((((....))))))..))))))........((((((.....))--)))).((((((((((((----((...(((........)))...))))))))).))))).... ( -36.90) >Bclau.0 1983210 97 + 4303871/40-160 UUCAGCAACCAGCCAA----CCAUGGUCAGGUGCUAAAUCCAGCAG------UUUA--UCUGCAAGAUAAGAG-A----AGCAUGCACG--UAAGCAACCCCUUCUUCU-AAUCUUG--- ...((((.((.((((.----...))))..)))))).......((((------....--.))))(((((.((((-(----((..(((...--...)))....)))).)))-.))))).--- ( -26.30) >Banth.0 320603 115 + 5227293/40-160 CUCAGCAACGGGUUUU-----UUUAAUACCGUGCUAACUCCAGCAAGCCAUAUAAAGGCUUGGAAGAUGAGAAGAUGUGACCGAGUACAUAUAAGUGCUCUCCUUCUUAUCUUUAUGGUU ...((((.(((.....-----.......)))))))....(((.((((((.......))))))((((((((((((....((..((((((......)))))))))))))))))))).))).. ( -38.90) >Bcere.0 361552 115 + 5224283/40-160 CUCAGCAACGGGUUUU-----UUUAAUACCGUGCUAACUCCAGCAAGCCUAUGAAAGGCUUGGAAGAUGAGAAGAUGUGAACGAGUACAUAUAAGUGCUCUCCUUCUUAUCUUUAUGGUU ...((((.(((.....-----.......)))))))....(((.(((((((.....)))))))((((((((((((..(....)((((((......))))))..)))))))))))).))).. ( -42.60) >consensus CUCAGCAACCAGCCAU_____UUUAAUAACGUGCUAACUCCAGCAAGCCUUAUAAA__CUUGGAAGAUAAGAAGA____AGCGAGUACAUAUAAGCACCCUCCUCCUCAUAAUCAUGGUU ...((((.(((((((........)))).)))))))........((((((.......)))))).....((((((((((.....((((((......)))))).......))))))))))... (-14.31 = -17.62 + 3.31)

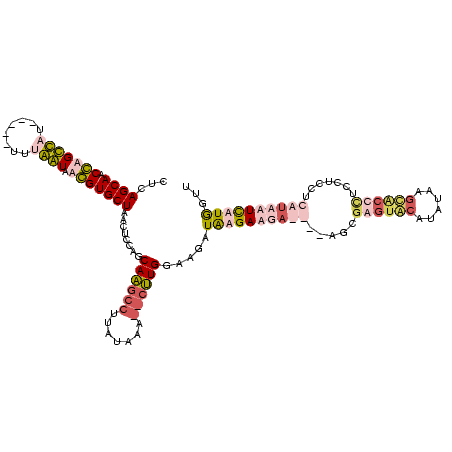
| Location | 1,699,955 – 1,700,069 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 58.20 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -17.52 |
| Energy contribution | -18.78 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 1699955 114 + 4202352/40-160 AUGUAAGAUUAAGAAGAAGUAGGUGUUGGAUCUCACUCGCU----UCCUCUUAUCUCUCAAG--CAAAUAAUGCUUGCAGGAAUUAGCACCUGACCACAGGAUCGUGGCUGGUUGCUGAG .........(((((.((((..((((........))))..))----)).)))))(((..((((--((.....))))))..))).((((((((.(.((((......))))).)).)))))). ( -39.10) >Bclau.0 1983210 97 + 4303871/40-160 ---CAAGAUU-AGAAGAAGGGGUUGCUUA--CGUGCAUGCU----U-CUCUUAUCUUGCAGA--UAAA------CUGCUGGAUUUAGCACCUGACCAUGG----UUGGCUGGUUGCUGAA ---.......-..((((.((((((((...--...))).)))----)-)))))((((.((((.--....------)))).))))((((((((.(.(((...----.)))).)).)))))). ( -29.40) >Banth.0 320603 115 + 5227293/40-160 AACCAUAAAGAUAAGAAGGAGAGCACUUAUAUGUACUCGGUCACAUCUUCUCAUCUUCCAAGCCUUUAUAUGGCUUGCUGGAGUUAGCACGGUAUUAAA-----AAAACCCGUUGCUGAG ..(((..(((((.((((((.(((.((......)).)))(....).)))))).))))).((((((.......)))))).)))..(((((((((.......-----.....))).)))))). ( -32.60) >Bcere.0 361552 115 + 5224283/40-160 AACCAUAAAGAUAAGAAGGAGAGCACUUAUAUGUACUCGUUCACAUCUUCUCAUCUUCCAAGCCUUUCAUAGGCUUGCUGGAGUUAGCACGGUAUUAAA-----AAAACCCGUUGCUGAG .......(((((..(((.(((.(((......))).))).)))..)))))....(((..(((((((.....)))))))..))).(((((((((.......-----.....))).)))))). ( -33.40) >consensus AACCAAAAAGAAAAAAAAGAGAGCACUUAUACGUACUCGCU____UCCUCUCAUCUUCCAAG__UAAAUAAGGCUUGCUGGAGUUAGCACCGGACCAAA_____AAAACCCGUUGCUGAG ....................((((((......))))))...............(((..((((((.......))))))..))).((((((((((.(((........))))))).)))))). (-17.52 = -18.78 + 1.25)

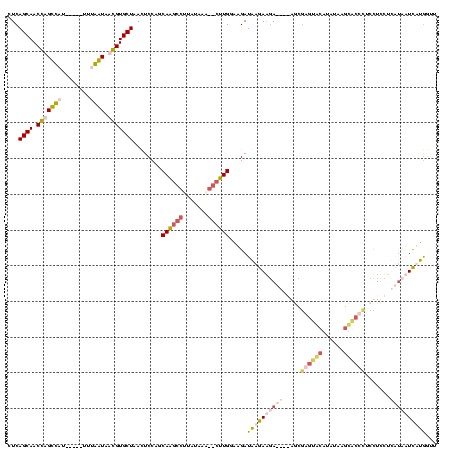
| Location | 1,699,955 – 1,700,069 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 59.10 |
| Mean single sequence MFE | -38.46 |
| Consensus MFE | -24.33 |
| Energy contribution | -25.09 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.41 |
| Mean z-score | -4.05 |
| Structure conservation index | 0.63 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 1699955 114 + 4202352/62-182 GUGGUCAGGUGCUAAUUCCUGCAAGCAUUAUUUG--CUUGAGAGAUAAGAGGA----AGCGAGUGAGAUCCAACACCUACUUCUUCUUAAUCUUACAUGACGAGAAGGUAGGUGUUUUUU (.((((...((((..(((((.((((((.....))--)))).........))))----)...)))).)))))(((((((((..((((((..((......)).))))))))))))))).... ( -34.10) >Bclau.0 1983210 95 + 4303871/62-182 AUGGUCAGGUGCUAAAUCCAGCAG------UUUA--UCUGCAAGAUAAGAG-A----AGCAUGCACG--UAAGCAACCCCUUCUUCU-AAUCUUG------GAGAAGG---GUUUUUUGU ...(((.((........)).((((------....--.))))..)))..(((-(----(((.(((...--...)))...((((((((.-......)------)))))))---))))))).. ( -26.50) >Banth.0 320603 120 + 5227293/62-182 UUAAUACCGUGCUAACUCCAGCAAGCCAUAUAAAGGCUUGGAAGAUGAGAAGAUGUGACCGAGUACAUAUAAGUGCUCUCCUUCUUAUCUUUAUGGUUGAUAAGAAGGAGAGCACUUUUU ........(((((...((((.((((((.......)))))).............)).))...)))))....(((((((((((((((((((.........)))))))))))))))))))... ( -45.04) >Bcere.0 361552 120 + 5224283/62-182 UUAAUACCGUGCUAACUCCAGCAAGCCUAUGAAAGGCUUGGAAGAUGAGAAGAUGUGAACGAGUACAUAUAAGUGCUCUCCUUCUUAUCUUUAUGGUUGAUAAGAAGGAGAGCACUUUUU ........(((((..(((...(((((((.....)))))))......)))....((....)))))))....(((((((((((((((((((.........)))))))))))))))))))... ( -48.20) >consensus UUAAUAACGUGCUAACUCCAGCAAGCCUUAUAAA__CUUGGAAGAUAAGAAGA____AGCGAGUACAUAUAAGCACCCUCCUCCUCAUAAUCAUGGUUGAUAAGAAGGAGAGCACUUUUU ........(((((...((...((((((.......))))))...))................)))))....(((((((((((((((((((.........)))))))))))))))))))... (-24.33 = -25.09 + 0.75)

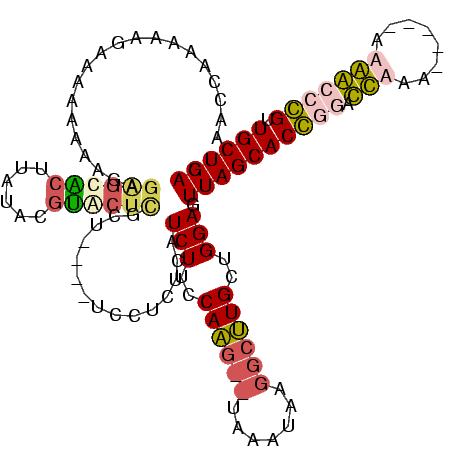
| Location | 1,699,955 – 1,700,069 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 59.10 |
| Mean single sequence MFE | -36.83 |
| Consensus MFE | -23.96 |
| Energy contribution | -25.77 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.32 |
| Mean z-score | -5.10 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 1699955 114 + 4202352/62-182 AAAAAACACCUACCUUCUCGUCAUGUAAGAUUAAGAAGAAGUAGGUGUUGGAUCUCACUCGCU----UCCUCUUAUCUCUCAAG--CAAAUAAUGCUUGCAGGAAUUAGCACCUGACCAC ....((((((((((((((.(((......)))..)))))..)))))))))((............----.............((((--((.....))))))((((........)))).)).. ( -29.50) >Bclau.0 1983210 95 + 4303871/62-182 ACAAAAAAC---CCUUCUC------CAAGAUU-AGAAGAAGGGGUUGCUUA--CGUGCAUGCU----U-CUCUUAUCUUGCAGA--UAAA------CUGCUGGAUUUAGCACCUGACCAU .........---((((((.------(......-.).))))))((((((...--...)).((((----.-.....((((.((((.--....------)))).))))..))))...)))).. ( -24.10) >Banth.0 320603 120 + 5227293/62-182 AAAAAGUGCUCUCCUUCUUAUCAACCAUAAAGAUAAGAAGGAGAGCACUUAUAUGUACUCGGUCACAUCUUCUCAUCUUCCAAGCCUUUAUAUGGCUUGCUGGAGUUAGCACGGUAUUAA ...(((((((((((((((((((.........)))))))))))))))))))....((((.((..............(((..((((((.......))))))..)))(....).))))))... ( -46.10) >Bcere.0 361552 120 + 5224283/62-182 AAAAAGUGCUCUCCUUCUUAUCAACCAUAAAGAUAAGAAGGAGAGCACUUAUAUGUACUCGUUCACAUCUUCUCAUCUUCCAAGCCUUUCAUAGGCUUGCUGGAGUUAGCACGGUAUUAA ...(((((((((((((((((((.........)))))))))))))))))))....((((.(((.............(((..(((((((.....)))))))..)))(....))))))))... ( -47.60) >consensus AAAAAAUACUCUCCUUCUCAUCAACCAAAAAGAAAAAAAAGAGAGCACUUAUACGUACUCGCU____UCCUCUCAUCUUCCAAG__UAAAUAAGGCUUGCUGGAGUUAGCACCGGACCAA ...(((((((((((((((((((.........))))))))))))))))))).........................(((..((((((.......))))))..)))................ (-23.96 = -25.77 + 1.81)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:26:54 2006