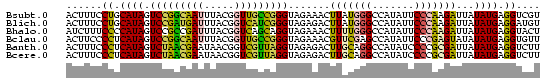
| Sequence ID | Bsubt.0 |
|---|---|
| Location | 26,692 – 697,803 |
| Length | 671111 |
| Max. P | 0.979774 |

| Location | 697,724 – 697,803 |
|---|---|
| Length | 79 |
| Sequences | 6 |
| Columns | 79 |
| Reading direction | forward |
| Mean pairwise identity | 78.90 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -23.66 |
| Energy contribution | -21.17 |
| Covariance contribution | -2.49 |
| Combinations/Pair | 1.68 |
| Mean z-score | -2.26 |
| Structure conservation index | 1.03 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 697724 79 + 4214630 ACGACCUCAUAUAAUCUUGGGAAUAUGGCCCAUAAGUUUCUACCCGGCAACCGUAAAUUGCCGGACUAUGCAGGAAAGU ....(((((((......((((.......))))...........(((((((.......)))))))..)))).)))..... ( -21.10) >Blich.0 697067 79 + 4222334 ACAUCCUCAUAUAAUCUUGGGAAUAUGGCCCAUAAGUCUCUACCCGAUGACCGUAAAUCAUCGGACUAUGCAGGAAAGU ...((((((((......((((.......))))...........(((((((.......)))))))..)))).)))).... ( -20.80) >Bhalo.0 676488 79 + 4202352 AGUACCUCAUAUAAUCUUGGGAAUAUGGCCCAAAAGUUUCUACCUGCUGACCGUAAAUCGGCGGACUAUGGGGAAAGAU ....(((((((.....(((((.......)))))..........(((((((.......)))))))..)))))))...... ( -22.40) >Bclau.0 1115678 79 + 4303871 AACACCUCAUAUAUACUCGGGAAUAUGGCUCGAACGUUUCUACCCGGCAACCGUAAAUUGCCGGACUAUGAGGGGAAGU ....(((((((.....(((((.......)))))..........(((((((.......)))))))..)))))))...... ( -24.90) >Banth.0 295344 79 + 5227293 AAGACCUCAUAUAAUCGCGGGGAUAUGGCCUGCAAGUCUCUACCUAACGACCGUUAUUCGUUAGACUAUGAGGGAAAGU ....(((((((.....(((((.......)))))..........(((((((.......)))))))..)))))))...... ( -24.40) >Bcere.0 336207 79 + 5224283 AAGACCUCAUAUAAUCGCGGGGAUAUGGCCUGCAAGUCUCUACCUAACGACCGUUAUUCGUUAGACUAUGAGGGAAAGU ....(((((((.....(((((.......)))))..........(((((((.......)))))))..)))))))...... ( -24.40) >consensus AAGACCUCAUAUAAUCUCGGGAAUAUGGCCCAAAAGUCUCUACCCGACGACCGUAAAUCGUCGGACUAUGAGGGAAAGU ....(((((((.....(((((.......)))))..........(((((((.......)))))))..)))))))...... (-23.66 = -21.17 + -2.49)

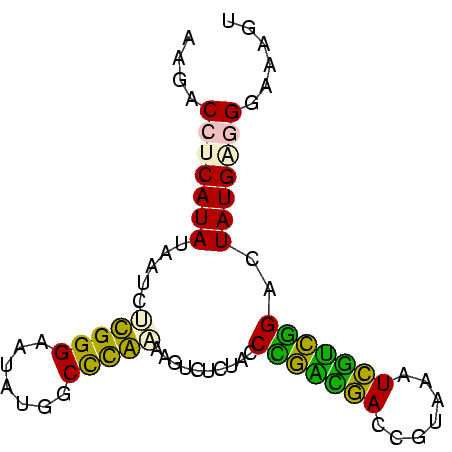
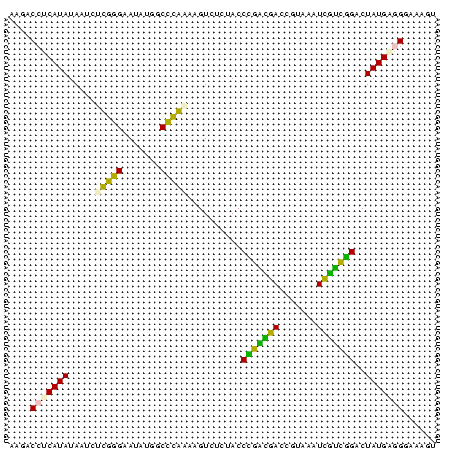
| Location | 697,724 – 697,803 |
|---|---|
| Length | 79 |
| Sequences | 6 |
| Columns | 79 |
| Reading direction | reverse |
| Mean pairwise identity | 78.90 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -19.17 |
| Energy contribution | -17.40 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 697724 79 + 4214630 ACUUUCCUGCAUAGUCCGGCAAUUUACGGUUGCCGGGUAGAAACUUAUGGGCCAUAUUCCCAAGAUUAUAUGAGGUCGU .....(((.((((.(((((((((.....)))))))))..........((((.......))))......))))))).... ( -22.70) >Blich.0 697067 79 + 4222334 ACUUUCCUGCAUAGUCCGAUGAUUUACGGUCAUCGGGUAGAGACUUAUGGGCCAUAUUCCCAAGAUUAUAUGAGGAUGU ....((((.((((.(((((((((.....)))))))))..........((((.......))))......))))))))... ( -23.00) >Bhalo.0 676488 79 + 4202352 AUCUUUCCCCAUAGUCCGCCGAUUUACGGUCAGCAGGUAGAAACUUUUGGGCCAUAUUCCCAAGAUUAUAUGAGGUACU .......((((((((..((((.....))))..))..........(((((((.......)))))))...)))).)).... ( -16.30) >Bclau.0 1115678 79 + 4303871 ACUUCCCCUCAUAGUCCGGCAAUUUACGGUUGCCGGGUAGAAACGUUCGAGCCAUAUUCCCGAGUAUAUAUGAGGUGUU ......(((((((.(((((((((.....))))))))).....(..((((...........))))..).))))))).... ( -23.50) >Banth.0 295344 79 + 5227293 ACUUUCCCUCAUAGUCUAACGAAUAACGGUCGUUAGGUAGAGACUUGCAGGCCAUAUCCCCGCGAUUAUAUGAGGUCUU ......(((((((.((((((((.......)))))))).......((((.((.......)).))))...))))))).... ( -20.70) >Bcere.0 336207 79 + 5224283 ACUUUCCCUCAUAGUCUAACGAAUAACGGUCGUUAGGUAGAGACUUGCAGGCCAUAUCCCCGCGAUUAUAUGAGGUCUU ......(((((((.((((((((.......)))))))).......((((.((.......)).))))...))))))).... ( -20.70) >consensus ACUUUCCCUCAUAGUCCGACGAUUUACGGUCGUCAGGUAGAAACUUACGGGCCAUAUUCCCAAGAUUAUAUGAGGUCUU ......((.((((.(((((((((.....))))))))).......(((((((.......)))))))...)))).)).... (-19.17 = -17.40 + -1.77)
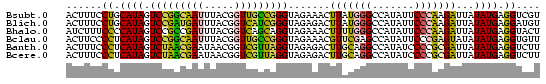
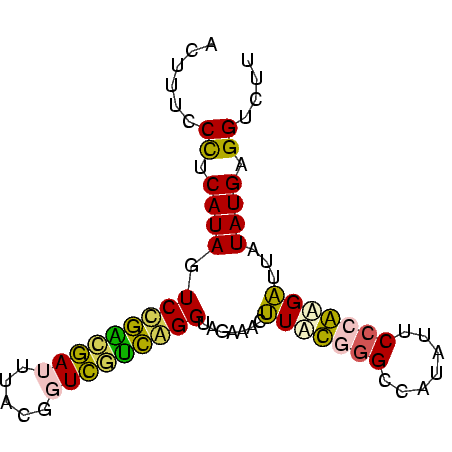
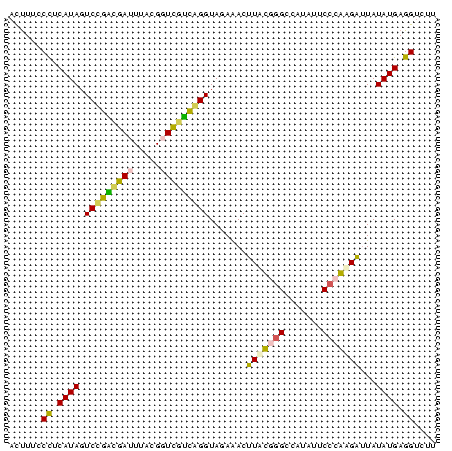
| Location | 697,724 – 697,803 |
|---|---|
| Length | 79 |
| Sequences | 6 |
| Columns | 79 |
| Reading direction | forward |
| Mean pairwise identity | 78.90 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -23.66 |
| Energy contribution | -21.17 |
| Covariance contribution | -2.49 |
| Combinations/Pair | 1.68 |
| Mean z-score | -2.26 |
| Structure conservation index | 1.03 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 697724 79 + 4214630/0-79 ACGACCUCAUAUAAUCUUGGGAAUAUGGCCCAUAAGUUUCUACCCGGCAACCGUAAAUUGCCGGACUAUGCAGGAAAGU ....(((((((......((((.......))))...........(((((((.......)))))))..)))).)))..... ( -21.10) >Blich.0 697067 79 + 4222334/0-79 ACAUCCUCAUAUAAUCUUGGGAAUAUGGCCCAUAAGUCUCUACCCGAUGACCGUAAAUCAUCGGACUAUGCAGGAAAGU ...((((((((......((((.......))))...........(((((((.......)))))))..)))).)))).... ( -20.80) >Bhalo.0 676488 79 + 4202352/0-79 AGUACCUCAUAUAAUCUUGGGAAUAUGGCCCAAAAGUUUCUACCUGCUGACCGUAAAUCGGCGGACUAUGGGGAAAGAU ....(((((((.....(((((.......)))))..........(((((((.......)))))))..)))))))...... ( -22.40) >Bclau.0 1115678 79 + 4303871/0-79 AACACCUCAUAUAUACUCGGGAAUAUGGCUCGAACGUUUCUACCCGGCAACCGUAAAUUGCCGGACUAUGAGGGGAAGU ....(((((((.....(((((.......)))))..........(((((((.......)))))))..)))))))...... ( -24.90) >Banth.0 295344 79 + 5227293/0-79 AAGACCUCAUAUAAUCGCGGGGAUAUGGCCUGCAAGUCUCUACCUAACGACCGUUAUUCGUUAGACUAUGAGGGAAAGU ....(((((((.....(((((.......)))))..........(((((((.......)))))))..)))))))...... ( -24.40) >Bcere.0 336207 79 + 5224283/0-79 AAGACCUCAUAUAAUCGCGGGGAUAUGGCCUGCAAGUCUCUACCUAACGACCGUUAUUCGUUAGACUAUGAGGGAAAGU ....(((((((.....(((((.......)))))..........(((((((.......)))))))..)))))))...... ( -24.40) >consensus AAGACCUCAUAUAAUCUCGGGAAUAUGGCCCAAAAGUCUCUACCCGACGACCGUAAAUCGUCGGACUAUGAGGGAAAGU ....(((((((.....(((((.......)))))..........(((((((.......)))))))..)))))))...... (-23.66 = -21.17 + -2.49)
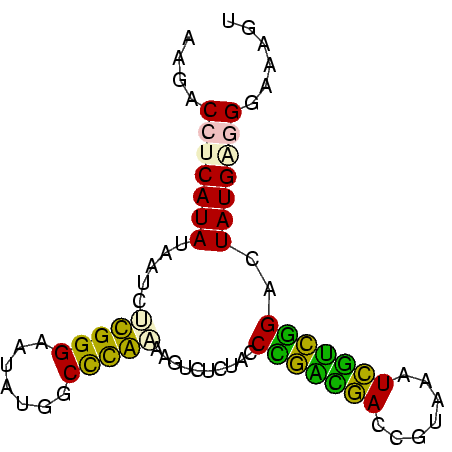
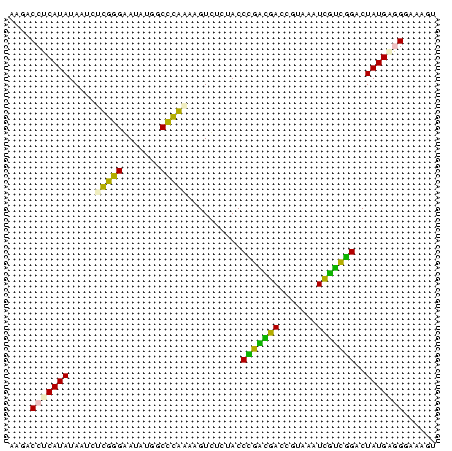
| Location | 697,724 – 697,803 |
|---|---|
| Length | 79 |
| Sequences | 6 |
| Columns | 79 |
| Reading direction | reverse |
| Mean pairwise identity | 78.90 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -19.17 |
| Energy contribution | -17.40 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 697724 79 + 4214630/0-79 ACUUUCCUGCAUAGUCCGGCAAUUUACGGUUGCCGGGUAGAAACUUAUGGGCCAUAUUCCCAAGAUUAUAUGAGGUCGU .....(((.((((.(((((((((.....)))))))))..........((((.......))))......))))))).... ( -22.70) >Blich.0 697067 79 + 4222334/0-79 ACUUUCCUGCAUAGUCCGAUGAUUUACGGUCAUCGGGUAGAGACUUAUGGGCCAUAUUCCCAAGAUUAUAUGAGGAUGU ....((((.((((.(((((((((.....)))))))))..........((((.......))))......))))))))... ( -23.00) >Bhalo.0 676488 79 + 4202352/0-79 AUCUUUCCCCAUAGUCCGCCGAUUUACGGUCAGCAGGUAGAAACUUUUGGGCCAUAUUCCCAAGAUUAUAUGAGGUACU .......((((((((..((((.....))))..))..........(((((((.......)))))))...)))).)).... ( -16.30) >Bclau.0 1115678 79 + 4303871/0-79 ACUUCCCCUCAUAGUCCGGCAAUUUACGGUUGCCGGGUAGAAACGUUCGAGCCAUAUUCCCGAGUAUAUAUGAGGUGUU ......(((((((.(((((((((.....))))))))).....(..((((...........))))..).))))))).... ( -23.50) >Banth.0 295344 79 + 5227293/0-79 ACUUUCCCUCAUAGUCUAACGAAUAACGGUCGUUAGGUAGAGACUUGCAGGCCAUAUCCCCGCGAUUAUAUGAGGUCUU ......(((((((.((((((((.......)))))))).......((((.((.......)).))))...))))))).... ( -20.70) >Bcere.0 336207 79 + 5224283/0-79 ACUUUCCCUCAUAGUCUAACGAAUAACGGUCGUUAGGUAGAGACUUGCAGGCCAUAUCCCCGCGAUUAUAUGAGGUCUU ......(((((((.((((((((.......)))))))).......((((.((.......)).))))...))))))).... ( -20.70) >consensus ACUUUCCCUCAUAGUCCGACGAUUUACGGUCGUCAGGUAGAAACUUACGGGCCAUAUUCCCAAGAUUAUAUGAGGUCUU ......((.((((.(((((((((.....))))))))).......(((((((.......)))))))...)))).)).... (-19.17 = -17.40 + -1.77)
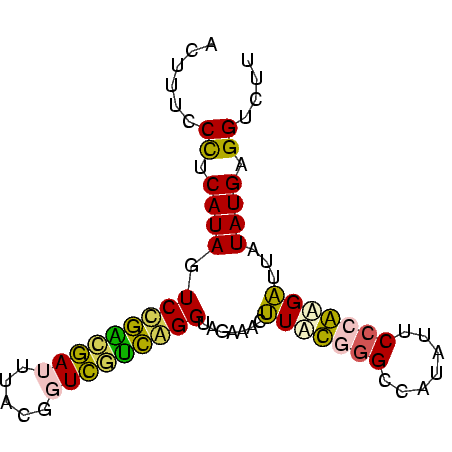
| Location | 26,692 – 26,731 |
|---|---|
| Length | 39 |
| Sequences | 5 |
| Columns | 43 |
| Reading direction | forward |
| Mean pairwise identity | 61.76 |
| Mean single sequence MFE | -15.16 |
| Consensus MFE | -16.62 |
| Energy contribution | -14.74 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.77 |
| Mean z-score | -5.76 |
| Structure conservation index | 1.10 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916338 |
| Prediction | RNA |
| WARNING | Sequence 1 too short. |
| WARNING | Sequence 2 too short. |
| WARNING | Sequence 2: Base composition out of range. |
| WARNING | Sequence 3 too short. |
| WARNING | Sequence 4 too short. |
| WARNING | Sequence 5 too short. |
Download alignment: ClustalW | MAF
>Bsubt.0 26692 39 + 4214630 AUCAAAUUUUCAGACUCACCUAAU---AU-UAGGUGAGUUUUU ...........(((((((((((..---..-))))))))))).. ( -14.20) >Blich.0 30958 42 + 4222334 AUCAAAUUAAGA-ACUCAUCUAUUUGAAUAUAGAUGAGUUUUU ........((((-((((((((((......)))))))))))))) ( -15.60) >Banth.0 25384 41 + 5227293 AUAUGUAUACAAAACUCACCUUAAU--ACUAAGGUGGGUUUUU ..........(((((((((((((..--..))))))))))))). ( -15.20) >Bcere.0 25385 41 + 5224283 AUAUGUAUACAAAACUCACCUUAAU--ACUAAGGUGGGUUUUU ..........(((((((((((((..--..))))))))))))). ( -15.20) >Bhalo.0 46755 42 + 4202352 ACUUAACCAUAAAAAGCAACUACAUU-CCGUAGUUGCUUUUUU ..........(((((((((((((...-..))))))))))))). ( -15.60) >consensus AUAUAAAUACAAAACUCACCUAAAU__ACUUAGGUGAGUUUUU ..........(((((((((((((......))))))))))))). (-16.62 = -14.74 + -1.88)
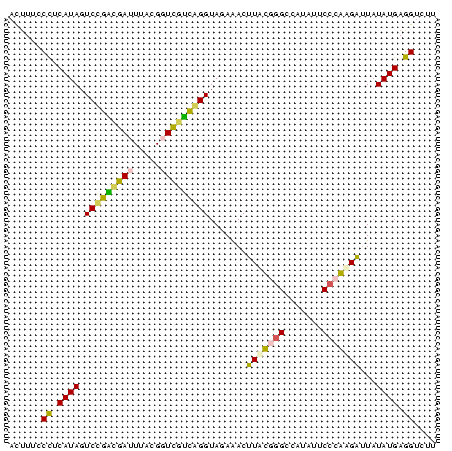
| Location | 26,692 – 26,731 |
|---|---|
| Length | 39 |
| Sequences | 5 |
| Columns | 43 |
| Reading direction | reverse |
| Mean pairwise identity | 61.76 |
| Mean single sequence MFE | -12.66 |
| Consensus MFE | -13.06 |
| Energy contribution | -12.54 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.54 |
| Mean z-score | -4.21 |
| Structure conservation index | 1.03 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901925 |
| Prediction | RNA |
| WARNING | Sequence 1 too short. |
| WARNING | Sequence 2 too short. |
| WARNING | Sequence 2: Base composition out of range. |
| WARNING | Sequence 3 too short. |
| WARNING | Sequence 4 too short. |
| WARNING | Sequence 5 too short. |
Download alignment: ClustalW | MAF
>Bsubt.0 26692 39 + 4214630 AAAAACUCACCUA-AU---AUUAGGUGAGUCUGAAAAUUUGAU ....(((((((((-..---..)))))))))............. ( -12.30) >Blich.0 30958 42 + 4222334 AAAAACUCAUCUAUAUUCAAAUAGAUGAGU-UCUUAAUUUGAU ...(((((((((((......))))))))))-)........... ( -12.70) >Banth.0 25384 41 + 5227293 AAAAACCCACCUUAGU--AUUAAGGUGAGUUUUGUAUACAUAU .(((((.(((((((..--..))))))).))))).......... ( -11.20) >Bcere.0 25385 41 + 5224283 AAAAACCCACCUUAGU--AUUAAGGUGAGUUUUGUAUACAUAU .(((((.(((((((..--..))))))).))))).......... ( -11.20) >Bhalo.0 46755 42 + 4202352 AAAAAAGCAACUACGG-AAUGUAGUUGCUUUUUAUGGUUAAGU .(((((((((((((..-...))))))))))))).......... ( -15.90) >consensus AAAAACCCACCUAAGU__AUUUAGGUGAGUUUUAUAAUUAGAU .(((((((((((((......))))))))))))).......... (-13.06 = -12.54 + -0.52)
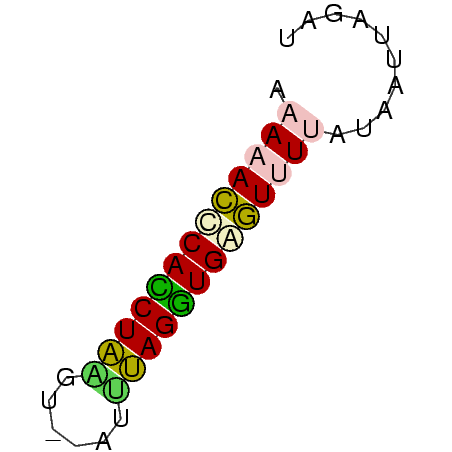
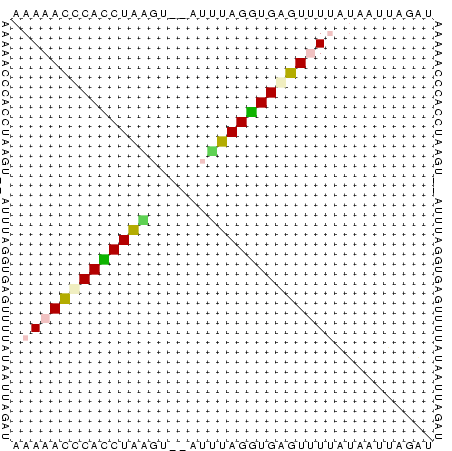
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:26:49 2006