| Sequence ID | Banth.0 |
|---|---|
| Location | 290,091 – 290,211 |
| Length | 120 |
| Max. P | 0.999994 |

| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.93 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -40.80 |
| Energy contribution | -39.64 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
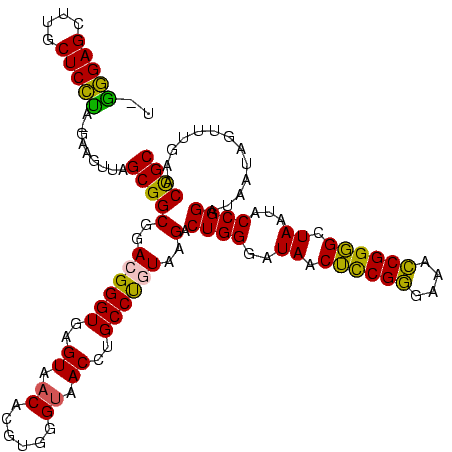
>Banth.0 290091 120 + 5227293/40-160 UUAAGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGC .((((((....))))))...........((((..((((..((.((......)).))..))))......((((..((.((((((....)))))).))...)))).............)))) ( -40.50) >Bcere.0 330810 120 + 5224283/40-160 UUAAGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGC .((((((....))))))...........((((..((((..((.((......)).))..))))......((((..((.((((((....)))))).))...)))).............)))) ( -40.50) >Bhalo.0 1017954 118 + 4202352/40-160 A-GGGAGCUUGCUCCUA-GAGGUUAGCGGCGAACGGGUGAGUAACACGUGGGCAACCUGCCUGUAAGACUGGGAUAACAUCGAGAAAUCGGUGCUAAUACCGGAUAAUAAAAAGAACUGC .-(((((....))))).-..((((.((((((.(((.((.....)).)))((....)))))).))....((((..((.((((((....)))))).))...))))...........)))).. ( -34.90) >Bsubt.0 160926 118 + 4214630/40-160 U-GGGAGCUUGCUCCCU-GAUGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGC .-(((((....))))).-.......(((((..((((((..((.((......)).))..))))))..).((((..((.((((((....)))))).))...)))).............)))) ( -44.90) >Blich.0 158327 118 + 4222334/40-160 C-GGGAGCUUGCUCCCU-UAGGUCAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAACCGC .-(((((....))))).-..(((((((((((.((((((..((.((......)).))..))))))....((((..((.((((((....)))))).))...)))).))).)).))).))).. ( -46.80) >consensus U_GGGAGCUUGCUCCUA_GAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAAUAGUUUGAACCGC ..(((((....))))).........(((((..((((((..((.((......)).))..))))))..).((((..((.((((((....)))))).))...)))).............)))) (-40.80 = -39.64 + -1.16)

| Location | 290,091 – 290,209 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.03 |
| Mean single sequence MFE | -45.62 |
| Consensus MFE | -41.72 |
| Energy contribution | -37.88 |
| Covariance contribution | -3.84 |
| Combinations/Pair | 1.49 |
| Mean z-score | -3.98 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.45 |
| SVM RNA-class probability | 0.999900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
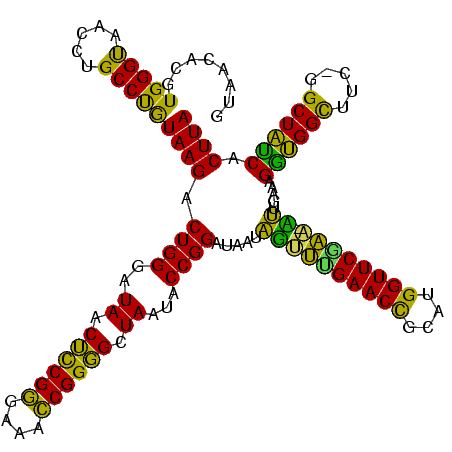
>Banth.0 290091 118 + 5227293/80-200 GUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAA-AUUGAAAGGCGGCUUC-GGCUGUCACUUA .......((((((.....))))))....((((..((.((((((....)))))).))...)))).....((((((((((....))))))))-)).....((((((...-.))))))..... ( -48.90) >Bcere.0 330810 118 + 5224283/80-200 GUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAA-AUUGAAAGGCGGCUUC-GGCUGUCACUUA .......((((((.....))))))....((((..((.((((((....)))))).))...)))).....((((((((((....))))))))-)).....((((((...-.))))))..... ( -48.90) >Bhalo.0 1017954 118 + 4202352/80-200 GUAACACGUGGGCAACCUGCCUGUAAGACUGGGAUAACAUCGAGAAAUCGGUGCUAAUACCGGAUAAUAAAAAGAACUGCAUGGUUCUUU-UUUGAAAGAUGGUUUC-GGCUAUCACUUA .......(..(((.....)))..)....((((..((.((((((....)))))).))...)))).....((((((((((....))))))))-)).....((((((...-.))))))..... ( -34.70) >Bsubt.0 160926 118 + 4214630/80-200 GUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGCAUGGUUCAAA-CAUAAAAGGUGGCUUC-GGCUACCACUUA ...(((.(..((...))..).)))....((((..((.((((((....)))))).))...)))).....((((((((((....))))))))-)).....((((((...-.))))))..... ( -50.70) >Blich.0 158327 120 + 4222334/80-200 GUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAACCGCAUGGUUCCAAUCAUAAAAGGUGGCUUUUAGCUACCACUUA ((((((.(..((...))..).)))....((((..((.((((((....)))))).))...)))).))).((((((((((....)))).)))))).....((((((.....))))))..... ( -44.90) >consensus GUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAAUAGUUUGAACCGCAUGGUUCGAA_AUUGAAAGGUGGCUUC_GGCUAUCACUUA ........(((((.....)))))((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))).)).....((((((.....)))))).)))) (-41.72 = -37.88 + -3.84)

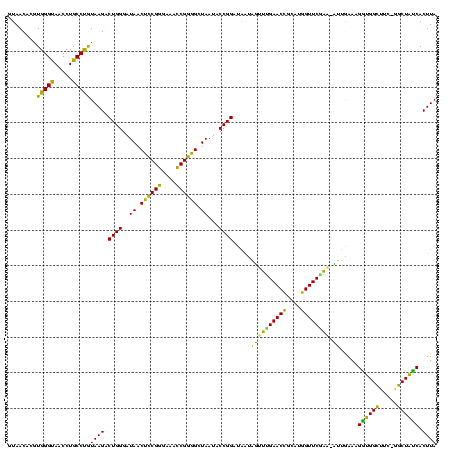
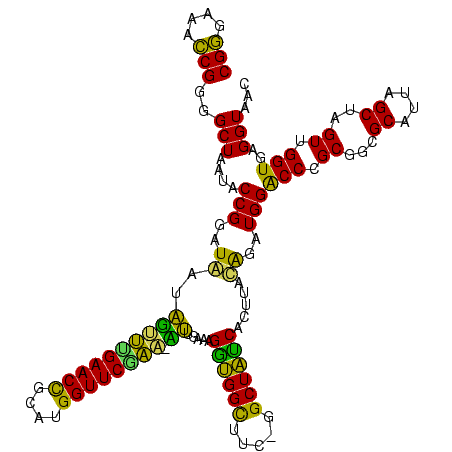
| Location | 290,091 – 290,209 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.52 |
| Mean single sequence MFE | -41.08 |
| Consensus MFE | -33.98 |
| Energy contribution | -30.74 |
| Covariance contribution | -3.24 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 118 + 5227293/120-240 CGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAA-AUUGAAAGGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAAC (((....)))((((((...(((......((((((((((....))))))))-)).....((((((...-.)))))).....))).))).)))((.((((............)))).))... ( -41.90) >Bcere.0 330810 118 + 5224283/120-240 CGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAA-AUUGAAAGGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAAC (((....)))((((((...(((......((((((((((....))))))))-)).....((((((...-.)))))).....))).))).)))((.((((............)))).))... ( -41.90) >Bhalo.0 1017954 118 + 4202352/120-240 CGAGAAAUCGGUGCUAAUACCGGAUAAUAAAAAGAACUGCAUGGUUCUUU-UUUGAAAGAUGGUUUC-GGCUAUCACUUACAGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUGGGGUAAC (((....))).((((..(((((......((((((((((....))))))))-)).....((((((...-.))))))(((..(.((((.(((...))).)))).)..)))))))).)))).. ( -33.40) >Bsubt.0 160926 118 + 4214630/120-240 CGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGCAUGGUUCAAA-CAUAAAAGGUGGCUUC-GGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAAU (((....)))..(((..((((((.((((((..((..((((..((((((..-..(((..((((((...-.))))))..)))....)))))).))))..)).)))))).)))))).)))... ( -46.60) >Blich.0 158327 120 + 4222334/120-240 CGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAACCGCAUGGUUCCAAUCAUAAAAGGUGGCUUUUAGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAAC (((....)))..(((..((((((..(((....((..((((..(((.(((.((.(((..((((((.....))))))..)))..)))))))).))))..))..)))...)))))).)))... ( -41.60) >consensus CGGGAAACCGGGGCUAAUACCGGAUAAUAGUUUGAACCGCAUGGUUCGAA_AUUGAAAGGUGGCUUC_GGCUAUCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAAC (((....)))..(((....(((..((..((((((((((....)))))))).)).....((((((.....)))))).....))..)))(((.((...((....))..)).)))..)))... (-33.98 = -30.74 + -3.24)

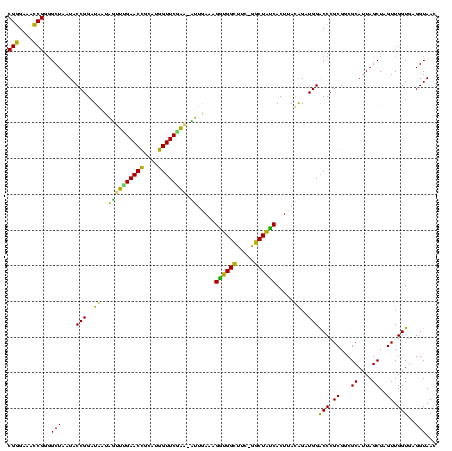
| Location | 290,091 – 290,209 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -46.50 |
| Consensus MFE | -41.34 |
| Energy contribution | -42.24 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 118 + 5227293/160-280 AUGGUUCGAA-AUUGAAAGGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGU ..((((((..-..(((..((((((...-.))))))..)))....))))))(((((((.....(((..((((((((.......)))))))))))..)))))))......((((....)))) ( -46.10) >Bcere.0 330810 118 + 5224283/160-280 AUGGUUCGAA-AUUGAAAGGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGU ..((((((..-..(((..((((((...-.))))))..)))....))))))(((((((.....(((..((((((((.......)))))))))))..)))))))......((((....)))) ( -46.10) >Bhalo.0 1017954 118 + 4202352/160-280 AUGGUUCUUU-UUUGAAAGAUGGUUUC-GGCUAUCACUUACAGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUGGGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGU ..(((((.((-(.(((..((((((...-.))))))..))).))).))))).((.(((((((.(((..((((((((.......)))))))))))...))))))).))..((((....)))) ( -42.70) >Bsubt.0 160926 118 + 4214630/160-280 AUGGUUCAAA-CAUAAAAGGUGGCUUC-GGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGU ..((((((..-..(((..((((((...-.))))))..)))....)))))).((.(((((((.(((..((((((((.......)))))))))))...))))))).))..((((....)))) ( -49.30) >Blich.0 158327 120 + 4222334/160-280 AUGGUUCCAAUCAUAAAAGGUGGCUUUUAGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGU ..((.((((.((.(((..((((((.....))))))..)))..)))))).))((.(((((((.(((..((((((((.......)))))))))))...))))))).))..((((....)))) ( -48.30) >consensus AUGGUUCGAA_AUUGAAAGGUGGCUUC_GGCUAUCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGU ..(((((......(((..((((((.....))))))..))).....))))).((..((((((.(((..((((((((.......)))))))))))...))))))..))..((((....)))) (-41.34 = -42.24 + 0.90)

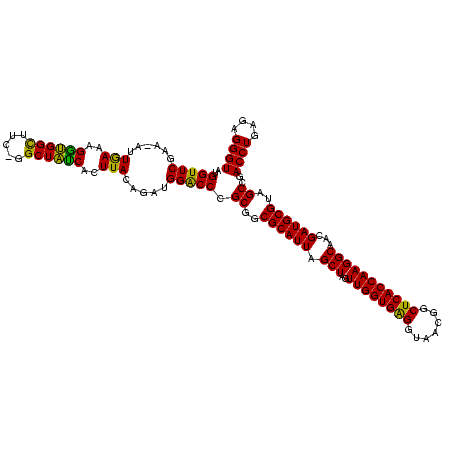

| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -48.80 |
| Consensus MFE | -43.80 |
| Energy contribution | -46.14 |
| Covariance contribution | 2.34 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/200-320 UGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUA .(((.......((((((.....(((..((((((((.......)))))))))))..))))))((.((((((((....)))...))))).)).(((((.(((.....))))))))..))).. ( -48.70) >Bcere.0 330810 120 + 5224283/200-320 UGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUA .(((.......((((((.....(((..((((((((.......)))))))))))..))))))((.((((((((....)))...))))).)).(((((.(((.....))))))))..))).. ( -48.70) >Bhalo.0 1017954 120 + 4202352/200-320 CAGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUGGGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUA ....((((((.((.(((((((.(((..((((((((.......)))))))))))...))))))).))..((((....))))....)))).))(((((.(((.....))))))))....... ( -50.60) >Bsubt.0 160926 119 + 4214630/200-320 CAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUC-UA .((((((.((.((.(((((((.(((..((((((((.......)))))))))))...))))))).))..((((....))))....)))))..(((((.(((.....))))))))..))-). ( -47.70) >Blich.0 158327 120 + 4222334/200-320 CAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUA .....(((......(((((((.(((..((((((((.......)))))))))))...))))))).((((((((....)))...)))))....(((((.(((.....))))))))..))).. ( -48.30) >consensus CAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUA ....(((.((.((..((((((.(((..((((((((.......)))))))))))...))))))..))..((((....))))....)))))..(((((.(((.....))))))))....... (-43.80 = -46.14 + 2.34)

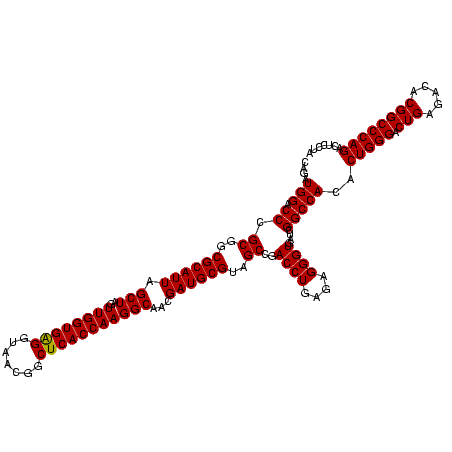

| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -49.82 |
| Consensus MFE | -45.98 |
| Energy contribution | -46.46 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.999272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/200-320 UAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCA ..(((...(((((((((((.((......((((((((((((.........))))))))))))...((((..((((((((.......)))))))).))))...)).))))).))))))))). ( -53.40) >Bcere.0 330810 120 + 5224283/200-320 UAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCA ..(((...(((((((((((.((......((((((((((((.........))))))))))))...((((..((((((((.......)))))))).))))...)).))))).))))))))). ( -53.40) >Bhalo.0 1017954 120 + 4202352/200-320 UAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCCCACCAACUAGCUAAUGCGCCGCGGGCCCAUCUG ..(((...((((((..............((((((((((((.........))))))))))))..(((.((.((((((...........))))))...((....)))).)))))))))))). ( -46.30) >Bsubt.0 160926 119 + 4214630/200-320 UA-GAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCAUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUG ((-((...((((((..............((((((((((((.........))))))))))))..(((.((.((((((((.......))))))))...((....)))).))))))))))))) ( -48.00) >Blich.0 158327 120 + 4222334/200-320 UAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUG ..(((...((((((..............((((((((((((.........))))))))))))..(((.((.((((((((.......))))))))...((....)))).)))))))))))). ( -48.00) >consensus UAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUG ..(((...((((((..............((((((((((((.........))))))))))))..(((.((.((((((((.......))))))))...((....)))).)))))))))))). (-45.98 = -46.46 + 0.48)

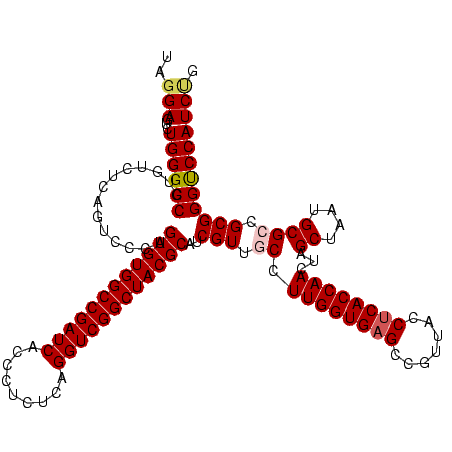
| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -44.34 |
| Consensus MFE | -43.66 |
| Energy contribution | -43.86 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.98 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/240-360 CUUUCGUCCAUUGCGGAAGAUUCCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCC ..((((.(((..(((((((((.(((.(((.((((.....)))).)))..)))....))))...)))..((((((((((((.........))))))))))))......))..))))))).. ( -44.70) >Bcere.0 330810 120 + 5224283/240-360 CUUUCGUCCAUUGCGGAAGAUUCCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCC ..((((.(((..(((((((((.(((.(((.((((.....)))).)))..)))....))))...)))..((((((((((((.........))))))))))))......))..))))))).. ( -44.70) >Bhalo.0 1017954 120 + 4202352/240-360 CUUUCGUCCAUUGCGGAAGAUUCCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCC ..((((.(((..(((((((((.(((.(((.((((.....)))).)))..)))....))))...)))..((((((((((((.........))))))))))))......))..))))))).. ( -45.00) >Bsubt.0 160926 119 + 4214630/240-360 CUUUCGUCCAUUGCGGAAGAUUCCCUACUGCUGCCUCCCGUA-GAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCC ..((((.(((..(((((((((.(((.(((.((((.....)))-))))..)))....))))...)))..((((((((((((.........))))))))))))......))..))))))).. ( -42.30) >Blich.0 158327 120 + 4222334/240-360 CUUUCGUCCAUUGCGGAAGAUUCCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCC ..((((.(((..(((((((((.(((.(((.((((.....)))).)))..)))....))))...)))..((((((((((((.........))))))))))))......))..))))))).. ( -45.00) >consensus CUUUCGUCCAUUGCGGAAGAUUCCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCC ..((((.(((..(((((((((.(((.(((.((((.....)))).)))..)))....))))...)))..((((((((((((.........))))))))))))......))..))))))).. (-43.66 = -43.86 + 0.20)


| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -41.42 |
| Consensus MFE | -38.16 |
| Energy contribution | -35.80 |
| Covariance contribution | -2.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.58 |
| SVM RNA-class probability | 0.999405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/360-480 UCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCA .............((.(((....))).))..(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..))))))). ( -41.80) >Bcere.0 330810 120 + 5224283/360-480 UCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCA .............((.(((....))).))..(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..))))))). ( -41.80) >Bhalo.0 1017954 119 + 4202352/360-480 UCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAA-AGGGCGGCACCUUGACGGUAUCUAACGAGAAAGCCA .............((.(((....))).))..(((((((..(((((....(((((....))))).....((((((((((((...-.)))))))).))))))))).((....))))))))). ( -44.30) >Bsubt.0 160926 120 + 4214630/360-480 UCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCA (((((((((((..((.(((....))).))..((((....)))).....))))))).((((((......((((((((((((.....)))))))).))))......))))))))))...... ( -43.90) >Blich.0 158327 120 + 4222334/360-480 UCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGCACCUUGACGGUACCUAACCAGAAAGCCA ((((((((((...((.(((....))).))..((((....))))......)))))).((((((......((((..((((((.....))))))...))))......))))))))))...... ( -35.30) >consensus UCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAAUAGGGCGGCACCUUGACGGUACCUAACCAGAAAGCCA .((.(((..((...))..))).)).......((((((((((.........)))...((((((......((((((((((((.....)))))))).))))......))))))..))))))). (-38.16 = -35.80 + -2.36)

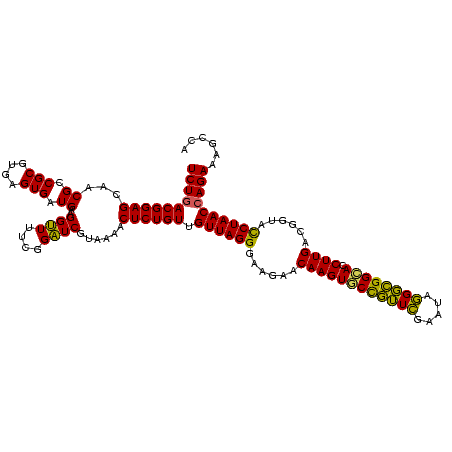
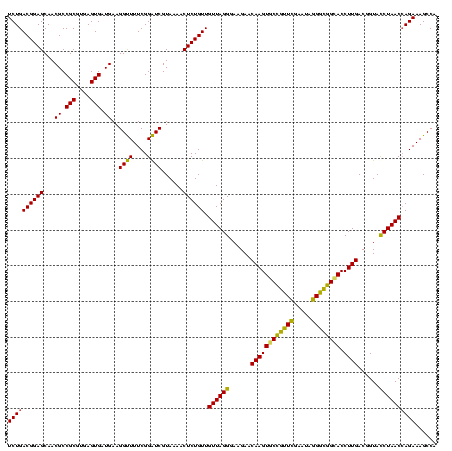
| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -41.26 |
| Consensus MFE | -41.08 |
| Energy contribution | -39.40 |
| Covariance contribution | -1.68 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.22 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.49 |
| SVM RNA-class probability | 0.999908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/400-520 GUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUG (((((.....((((..((((((......((((((((((((.....)))))))).))))......)))))))))).....)))))...(((((((((.((....)))))...))))))... ( -42.50) >Bcere.0 330810 120 + 5224283/400-520 GUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUG (((((.....((((..((((((......((((((((((((.....)))))))).))))......)))))))))).....)))))...(((((((((.((....)))))...))))))... ( -42.50) >Bhalo.0 1017954 119 + 4202352/400-520 AUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAA-AGGGCGGCACCUUGACGGUAUCUAACGAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUG (((((....(((((....))))).....((((((((((((...-.)))))))).))))))))).((....))...(((....)))..(((((((((.((....)))))...))))))... ( -43.50) >Bsubt.0 160926 120 + 4214630/400-520 AUCGUAAAGCUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUG ..........((((..((((((......((((((((((((.....)))))))).))))......)))))))))).(((....)))..(((((((((.((....)))))...))))))... ( -41.00) >Blich.0 158327 120 + 4222334/400-520 AUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUG ..........((((..((((((......((((..((((((.....))))))...))))......)))))))))).(((....)))..(((((((((.((....)))))...))))))... ( -36.80) >consensus AUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAAUAGGGCGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUG ..........((((..((((((......((((((((((((.....)))))))).))))......)))))))))).(((....)))..(((((((((.((....)))))...))))))... (-41.08 = -39.40 + -1.68)

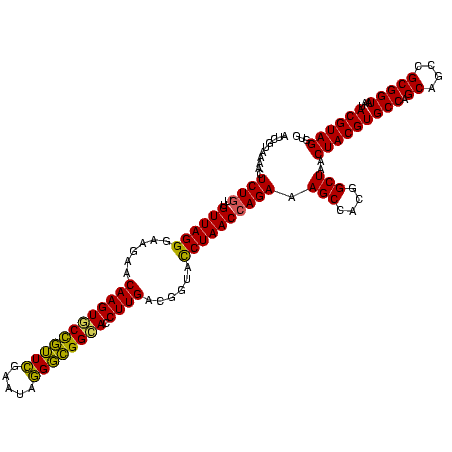
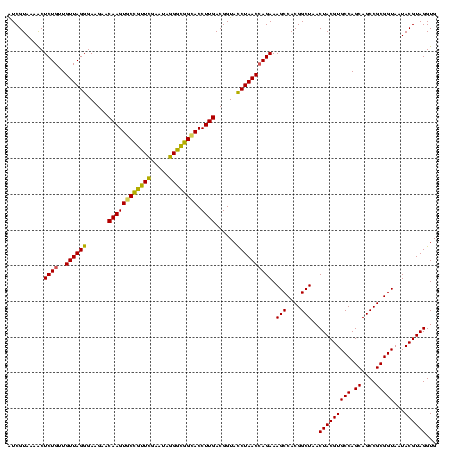
| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -38.47 |
| Consensus MFE | -37.72 |
| Energy contribution | -37.64 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/400-520 CACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCAGCUUAUUCAACUAGCACUUGUUCUUCCCUAACAACAGAGUUUUACGAC ......((((...((((((((((((.....))).)))))))))..(((((((((((........((((((.((.........)).)))))).......))))))..)))))...)))).. ( -37.56) >Bcere.0 330810 120 + 5224283/400-520 CACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCAGCUUAUUCAACUAGCACUUGUUCUUCCCUAACAACAGAGUUUUACGAC ......((((...((((((((((((.....))).)))))))))..(((((((((((........((((((.((.........)).)))))).......))))))..)))))...)))).. ( -37.56) >Bhalo.0 1017954 119 + 4202352/400-520 CACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUCGUUAGAUACCGUCAAGGUGCCGCCCU-UUCGAACGGCACUUGUUCUUCCCUAACAACAGAGUUUUACGAU ......((((...((((((((((((.....))).)))))))))((((...(((((.........((((((((..(.-...)..))))))))........)))))...))))...)))).. ( -36.43) >Bsubt.0 160926 120 + 4214630/400-520 CACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGUACUUGUUCUUCCCUAACAACAGAGCUUUACGAU ......((((...((((((((((((.....))).)))))))))..(((((((((((........((((((((..(.....)..)))))))).......))))))..)))))...)))).. ( -40.36) >Blich.0 158327 120 + 4222334/400-520 CACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCGCCCUAUUCGAACGGUACUUGUUCUUCCCUAACAACAGAGUUUUACGAU ......((((...((((((((((((.....))).)))))))))..(((((((((((........((((((((..(.....)..)))))))).......))))))..)))))...)))).. ( -40.46) >consensus CACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCGCCCUAUUCGAACGGCACUUGUUCUUCCCUAACAACAGAGUUUUACGAU ......((((...((((((((((((.....))).)))))))))..(((((((((((........((((((((...........)))))))).......))))))..)))))...)))).. (-37.72 = -37.64 + -0.08)

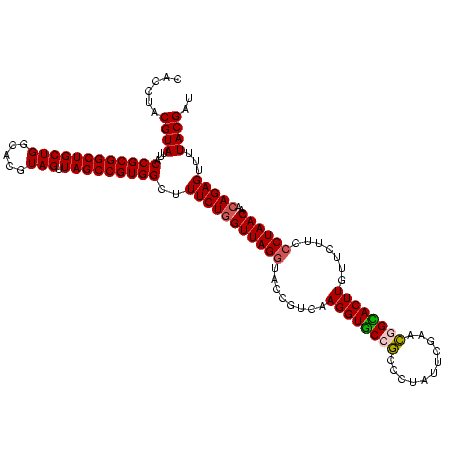
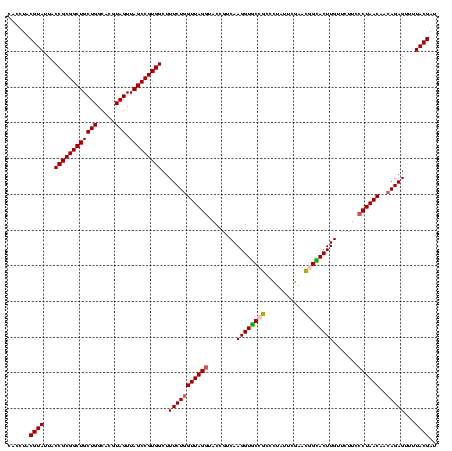
| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -26.59 |
| Consensus MFE | -23.44 |
| Energy contribution | -24.00 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/560-680 CUACGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAGAAACCACC .......((((...((.......((((.......))))......))....((((((....(((......)))..(((((((.....)))))))..........))))))..))))..... ( -24.02) >Bcere.0 330810 120 + 5224283/560-680 CUACGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAGAAACCACC .......((((...((.......((((.......))))......))....((((((....(((......)))..(((((((.....)))))))..........))))))..))))..... ( -24.02) >Bhalo.0 1017954 120 + 4202352/560-680 CUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGUACUCAAGUCUCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAGAGACCGCC ....((...............((((....))))...................(((((..(((((....(((...(((((((.....)))))))...)))....)))))...))))).)). ( -31.90) >Bsubt.0 160926 120 + 4214630/560-680 CUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUUCCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAGAAACCGCC ....((.((((..........((((....))))........((((.....((((.((((.((((.....((((......)))))))).))))))))......)))).....))))..)). ( -26.50) >Blich.0 158327 120 + 4222334/560-680 CUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUUCCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAGAAACCGCC ....((.((((..........((((....))))........((((.....((((.((((.((((.....((((......)))))))).))))))))......)))).....))))..)). ( -26.50) >consensus CUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAGAAACCGCC ....((........)).....((((....))))...................(((((..(((((....(((...(((((((.....)))))))...)))....)))))...))))).... (-23.44 = -24.00 + 0.56)

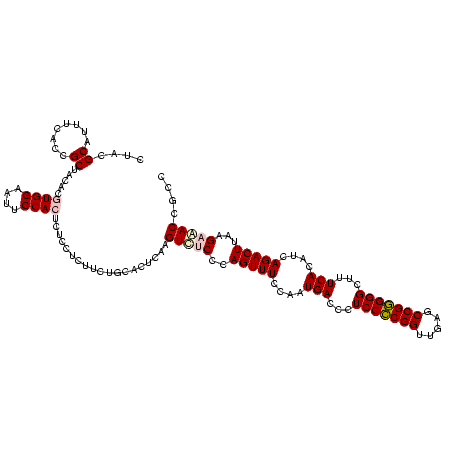
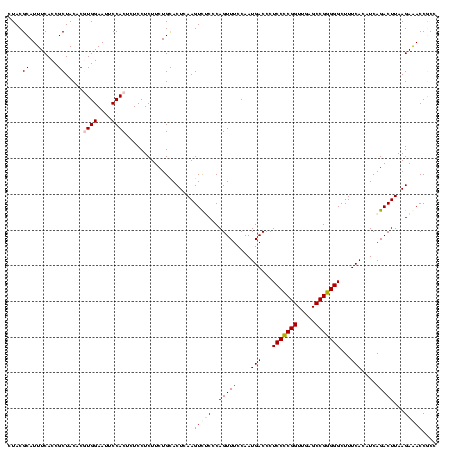
| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -36.86 |
| Energy contribution | -35.90 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.39 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.64 |
| SVM RNA-class probability | 0.999933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/600-720 CGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGU ..(.(((..((...(....)...))..))).)...(((((.......(((...((((((((.(.(((......)))....).))))))))...))).(((.((....)).)))))))).. ( -34.50) >Bcere.0 330810 120 + 5224283/600-720 CGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGU ..(.(((..((...(....)...))..))).)...(((((.......(((...((((((((.(.(((......)))....).))))))))...))).(((.((....)).)))))))).. ( -34.50) >Bhalo.0 1017954 120 + 4202352/600-720 CGGGGAGGGUCAUUGGAAACUGGGAGACUUGAGUACAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGU (((((((...((((((..(((((....))..))).............((....((((((((.(.(((......)))....).))))))))..)).))))))((....))..))))))).. ( -38.70) >Bsubt.0 160926 120 + 4214630/600-720 CGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGU ...((((((((.......(((((...((((.....(....)....))))(...((((((((.(.(((......)))....).))))))))...).))))).((....))))))))))... ( -37.90) >Blich.0 158327 120 + 4222334/600-720 CGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGU ...((((((((.......(((((...((((.....(....)....))))(...((((((((.(.(((......)))....).))))))))...).))))).((....))))))))))... ( -37.90) >consensus CGGGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGU ...((((((((.......(((((...((((.....(....)....))))(...((((((((.(.(((......)))....).))))))))...).))))).((....))))))))))... (-36.86 = -35.90 + -0.96)

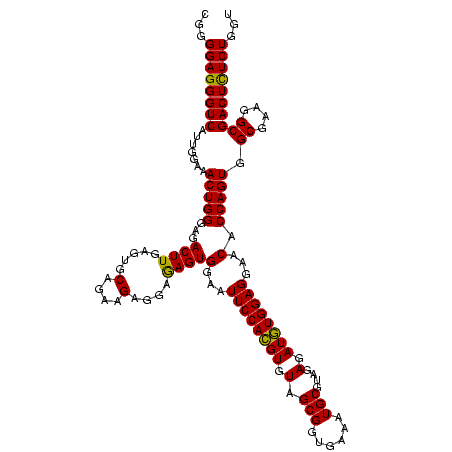
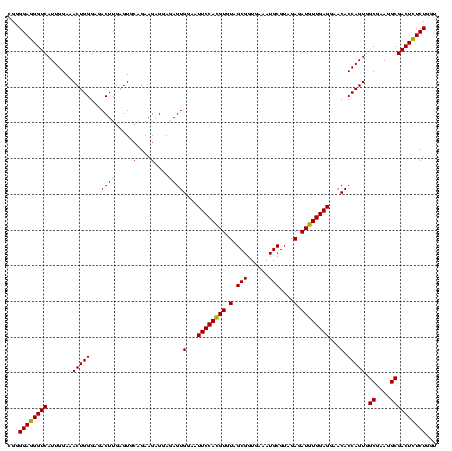
| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -39.88 |
| Consensus MFE | -36.62 |
| Energy contribution | -36.02 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/640-760 GAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAAC .......(((...((((((((.(.(((......)))....).))))))))...)))(((((((....))((((....)))).........)))))..((((....))))........... ( -38.20) >Bcere.0 330810 120 + 5224283/640-760 GAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAAC .......(((...((((((((.(.(((......)))....).))))))))...)))(((((((....))((((....)))).........)))))..((((....))))........... ( -38.20) >Bhalo.0 1017954 120 + 4202352/640-760 GAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAAC ..((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).))))))).(((.......)))(((...((((....))))....))).... ( -42.60) >Bsubt.0 160926 120 + 4214630/640-760 GAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGAGCGAAAACGUGGGGAGCGAAC ..((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))(.(((.((.....((((....))))......)).))).).... ( -37.80) >Blich.0 158327 120 + 4222334/640-760 GAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCGAAC ..((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).))))))).(((.......)))(((...((((....))))....))).... ( -42.60) >consensus GAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAAC ..((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).))))))).((.(((..((.(((((..........))))).))..))).)) (-36.62 = -36.02 + -0.60)

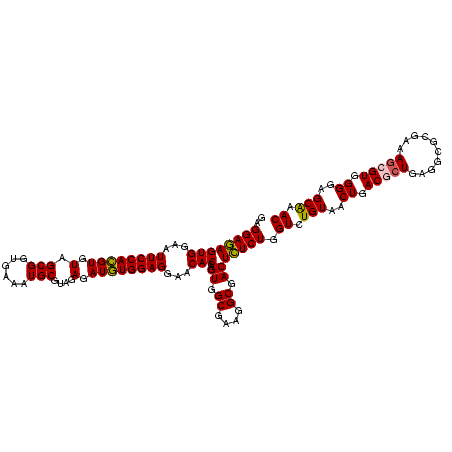
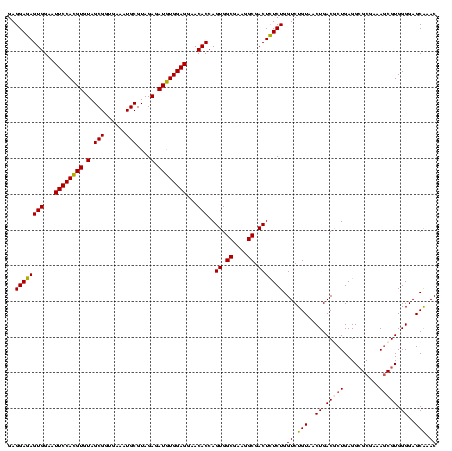
| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -41.42 |
| Consensus MFE | -39.70 |
| Energy contribution | -38.90 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.750999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/720-840 CUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGU ..(((.((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))(((((....)))))....)))...... ( -40.90) >Bcere.0 330810 120 + 5224283/720-840 CUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGU ..(((.((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))(((((....)))))....)))...... ( -40.90) >Bhalo.0 1017954 120 + 4202352/720-840 CUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGACGCCCUUAGUGCCGAAGU .....(((.(((....(((((....))(((((..((...((((.........))))))..)))))))).....)).).)))(((.((..((((((((......))))))))..))..))) ( -41.90) >Bsubt.0 160926 120 + 4214630/720-840 CUGUAACUGACGCUGAGGAGCGAAAACGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGC (((((.((((((((....((((....(((((...((...((((.........))))...))))))).((....)).....)))).))))))))(((((....))))).......))))). ( -39.30) >Blich.0 158327 120 + 4222334/720-840 CUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGCAGC (((((.((((((((..(((((....((((((...((...((((.........))))...))))))))((....))....))))).))))))))(((((....))))).......))))). ( -44.10) >consensus CUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGU ..(((.((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))((((......))))....)))...... (-39.70 = -38.90 + -0.80)


| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -41.38 |
| Consensus MFE | -41.04 |
| Energy contribution | -39.64 |
| Covariance contribution | -1.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/760-880 AGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAG .((((....(((....)))))))..(((((....((((.(((((.((..(((((((((....)))))))))..))..)))..)).))))....(((((......))))))))))...... ( -40.80) >Bcere.0 330810 120 + 5224283/760-880 AGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAG .((((....(((....)))))))..(((((....((((.(((((.((..(((((((((....)))))))))..))..)))..)).))))....(((((......))))))))))...... ( -40.80) >Bhalo.0 1017954 120 + 4202352/760-880 AGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGACGCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAG .((.((..((.(((((((.........((....))..(((((((.((..((((((((......))))))))..))...((....))....))))))).))).)))).))..))))..... ( -38.30) >Bsubt.0 160926 120 + 4214630/760-880 AGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAG ..((((..((.(((((((.........((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))).)))).))..))))..... ( -44.40) >Blich.0 158327 120 + 4222334/760-880 AGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGCAGCAAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAG .((((....(((....))))))).((((((....((((..((((.((..(((((((((....)))))))))..))..))))....))))....(((((......))))))))).)).... ( -42.60) >consensus AGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAG .((((....(((....)))))))..(((((....((((.(((((.((..((((((((......))))))))..))..)))..)).))))....(((((......))))))))))...... (-41.04 = -39.64 + -1.40)

| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -49.40 |
| Consensus MFE | -50.16 |
| Energy contribution | -48.20 |
| Covariance contribution | -1.96 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.79 |
| Structure conservation index | 1.02 |
| SVM decision value | 5.81 |
| SVM RNA-class probability | 0.999994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/800-920 UGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGG .(((.((..(((((((((....)))))))))..))..)))...........((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))) ( -49.00) >Bcere.0 330810 120 + 5224283/800-920 UGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGG .(((.((..(((((((((....)))))))))..))..)))...........((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))) ( -49.00) >Bhalo.0 1017954 120 + 4202352/800-920 UGCUAGGUGUUAGGGGUUUCGACGCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCAG ((((.((..((((((((......))))))))..))................((.((((((((...((((.(((((....)))))..))))..)))......))))))).......)))). ( -45.60) >Bsubt.0 160926 120 + 4214630/800-920 UGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGG (((..((..(((((((((....)))))))))..)))))((....)).....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))) ( -52.60) >Blich.0 158327 120 + 4222334/800-920 UGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGCAGCAAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGG ((((.((..(((((((((....)))))))))..))..))))..........((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))) ( -50.80) >consensus UGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGG .(((.((..((((((((......))))))))..))..)))...........((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))) (-50.16 = -48.20 + -1.96)

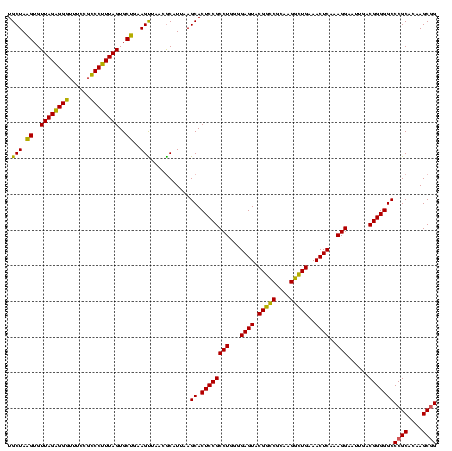
| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -40.14 |
| Consensus MFE | -39.58 |
| Energy contribution | -38.62 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/840-960 UAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUU ....((.....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))....))....(((((..((((........))))))))).. ( -41.80) >Bcere.0 330810 120 + 5224283/840-960 UAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUU ....((.....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))....))....(((((..((((........))))))))).. ( -41.80) >Bhalo.0 1017954 120 + 4202352/840-960 UAACACAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCAGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUU ....((((......((((((((...((((.(((((....)))))..))))..)))......)))))((((((.......)))).))))))(((((..((((........))))))))).. ( -37.70) >Bsubt.0 160926 120 + 4214630/840-960 UAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUU ....((.....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))....))....(((((..((((........))))))))).. ( -39.70) >Blich.0 158327 120 + 4222334/840-960 AAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUU ....((.....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))....))....(((((..((((........))))))))).. ( -39.70) >consensus UAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUU ....((((......((((((((...((((.(((((....)))))..))))..)))......)))))((((((.......)))).))))))(((((..((((........))))))))).. (-39.58 = -38.62 + -0.96)


| Location | 290,091 – 290,209 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -37.08 |
| Energy contribution | -38.96 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 118 + 5227293/920-1040 UGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAGGUCUUGACAUCCUCUGACAACCCUAGAGAUAGGGCUU-CUCCUUCGGGAG-CAGAGUGACAGGUGGUGCAUGGUU ..((.((((((((((..((((........)))))))))..((((...((((...(((((((....(((((....)))))...-((((....))))-))))).))))))))))))))).)) ( -38.90) >Bcere.0 330810 118 + 5224283/920-1040 UGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAGGUCUUGACAUCCUCUGACAACCCUAGAGAUAGGGCUU-CUCCUUCGGGAG-CAGAGUGACAGGUGGUGCAUGGUU ..((.((((((((((..((((........)))))))))..((((...((((...(((((((....(((((....)))))...-((((....))))-))))).))))))))))))))).)) ( -38.90) >Bhalo.0 1017954 120 + 4202352/920-1040 UGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAGGUCUUGACAUCCUUUGACCACCCUAGAGAUAGGGCUUUCCCCUUCGGGGGACAAAGUGACAGGUGGUGCAUGGUU ..((.((((((((((..((((........)))))))))..((((...((((...(((((((....(((((....)))))...(((((....)))))))))).))))))))))))))).)) ( -40.70) >Bsubt.0 160926 118 + 4214630/920-1040 UGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAGGUCUUGACAUCCUCUGACAAUCCUAGAGAUAGGACGU-CCCCUUCGGGGG-CAGAGUGACAGGUGGUGCAUGGUU ..((.((((((((((..((((........)))))))))..((((...((((...(((((((....(((((....)))))...-((((....))))-))))).))))))))))))))).)) ( -39.50) >Blich.0 158327 118 + 4222334/920-1040 UGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAGGUCUUGACAUCCUCUGACAACCCUAGAGAUAGGGCUU-CCCCUUCGGGGG-CAGAGUGACAGGUGGUGCAUGGUU ..((.((((((((((..((((........)))))))))..((((...((((...(((((((....(((((....)))))...-((((....))))-))))).))))))))))))))).)) ( -41.00) >consensus UGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAGGUCUUGACAUCCUCUGACAACCCUAGAGAUAGGGCUU_CCCCUUCGGGGG_CAGAGUGACAGGUGGUGCAUGGUU ..((.((((((((((..((((........)))))))))..((((...((((...(((((((....(((((....)))))....((((....)))).))))).))))))))))))))).)) (-37.08 = -38.96 + 1.88)

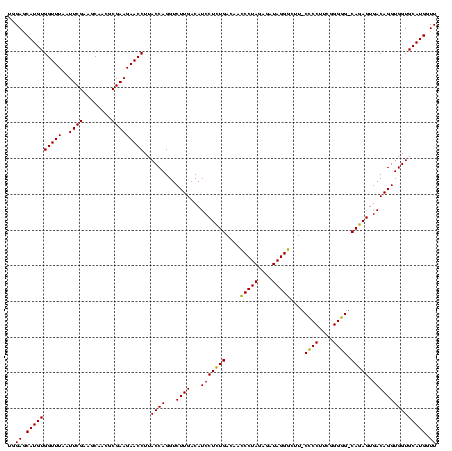
| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.67 |
| Mean single sequence MFE | -39.30 |
| Consensus MFE | -40.92 |
| Energy contribution | -37.08 |
| Covariance contribution | -3.84 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.71 |
| Structure conservation index | 1.04 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/1200-1320 GCUACACACGUGCUACAAUGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAU (((.((..((.(((.....(((((((.....(((..((...(((....)))...))....))).....)))))))..))).))...(((((((........))))))))).)))...... ( -35.10) >Bcere.0 330810 120 + 5224283/1200-1320 GCUACACACGUGCUACAAUGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAU (((.((..((.(((.....(((((((.....(((..((...(((....)))...))....))).....)))))))..))).))...(((((((........))))))))).)))...... ( -35.10) >Bhalo.0 1017954 120 + 4202352/1200-1320 GCUACACACGUGCUACAAUGGAUGGUACAAAGGGUUGCGAAGCCGCGAGGUGAAGCCAAUCCCAGAAAGCCAUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAAGCCGGAAU ((.((....))))......(((((((.....(((((((...(((....)))...).))))))......)))))))....(((((..(((((((........))))))).....))))).. ( -42.00) >Bsubt.0 160926 120 + 4214630/1200-1320 GCUACACACGUGCUACAAUGGACAGAACAAAGGGCAGCGAAACCGCGAGGUUAAGCCAAUCCCACAAAUCUGUUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGCUGGAAU (((.(((.((.(((.....(((((((.....(((..((..((((....))))..))....))).....)))))))..))).))....((((((........))))))))).)))...... ( -41.20) >Blich.0 158327 120 + 4222334/1200-1320 GCUACACACGUGCUACAAUGGGCAGAACAAAGGGCAGCGAAGCCGCGAGGCUAAGCCAAUCCCACAAAUCUGUUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGCUGGAAU (((.(((.((.(((.....(((((((.....(((..((..((((....))))..))....))).....)))))))..))).))....((((((........))))))))).)))...... ( -43.10) >consensus GCUACACACGUGCUACAAUGGACGGUACAAAGGGCUGCGAAACCGCGAGGUGAAGCCAAUCCCACAAAACCGUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAAGCUGGAAU (((.((..((.(((.....(((((((.....(((..((...(((....)))...))....))).....)))))))..))).))...(((((((........))))))))).)))...... (-40.92 = -37.08 + -3.84)

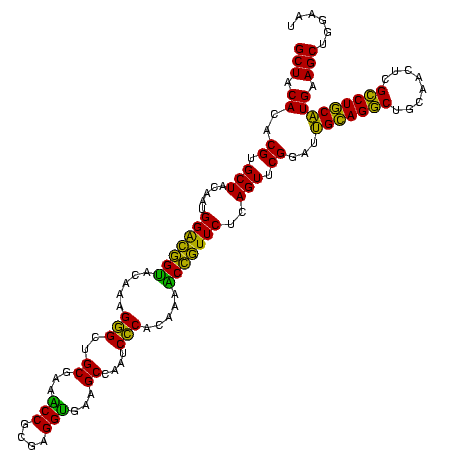
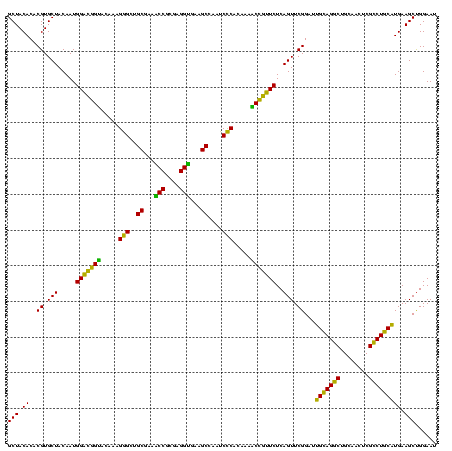
| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -40.68 |
| Consensus MFE | -38.92 |
| Energy contribution | -37.48 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/1240-1360 AACGUAUUCACCGCGGCAUGCUGAUCCGCGAUUACUAGCGAUUCCAGCUUCAUGUAGGCGAGUUGCAGCCUACAAUCCGAACUGAGAACGGUUUUAUGAGAUUAGCUCCACCUCGCGGUC .........(((((((...((((((((((........))).((((((.(((.(((((((........)))))))....)))))).)))...........)))))))......))))))). ( -38.70) >Bcere.0 330810 120 + 5224283/1240-1360 AACGUAUUCACCGCGGCAUGCUGAUCCGCGAUUACUAGCGAUUCCAGCUUCAUGUAGGCGAGUUGCAGCCUACAAUCCGAACUGAGAACGGUUUUAUGAGAUUAGCUCCACCUCGCGGUC .........(((((((...((((((((((........))).((((((.(((.(((((((........)))))))....)))))).)))...........)))))))......))))))). ( -38.70) >Bhalo.0 1017954 120 + 4202352/1240-1360 AACGUAUUCACCGCGGCAUGCUGAUCCGCGAUUACUAGCAAUUCCGGCUUCAUGCAGGCGAGUUGCAGCCUGCAAUCCGAACUGAGAAUGGCUUUCUGGGAUUGGCUUCACCUCGCGGCU ..........((((((...((..((((..((.....(((.(((((((.(((.(((((((........)))))))....)))))).)))).))).))..))))..))......)))))).. ( -42.90) >Bsubt.0 160926 120 + 4214630/1240-1360 AACGUAUUCACCGCGGCAUGCUGAUCCGCGAUUACUAGCGAUUCCAGCUUCACGCAGUCGAGUUGCAGACUGCGAUCCGAACUGAGAACAGAUUUGUGGGAUUGGCUUAACCUCGCGGUU .........(((((((...((..((((((........))(.((((((.(((.(((((((........)))))))....)))))).)))).........))))..))......))))))). ( -41.00) >Blich.0 158327 120 + 4222334/1240-1360 AACGUAUUCACCGCGGCAUGCUGAUCCGCGAUUACUAGCGAUUCCAGCUUCACGCAGUCGAGUUGCAGACUGCGAUCCGAACUGAGAACAGAUUUGUGGGAUUGGCUUAGCCUCGCGGCU ..........(((((((..((..((((((........))(.((((((.(((.(((((((........)))))))....)))))).)))).........))))..))...)))..)))).. ( -42.10) >consensus AACGUAUUCACCGCGGCAUGCUGAUCCGCGAUUACUAGCGAUUCCAGCUUCAUGCAGGCGAGUUGCAGCCUGCAAUCCGAACUGAGAACGGAUUUAUGGGAUUGGCUUCACCUCGCGGUU ..........((((((...((((((((((........))..((((((.(((.(((((((........)))))))....)))))).)))..........))))))))......)))))).. (-38.92 = -37.48 + -1.44)

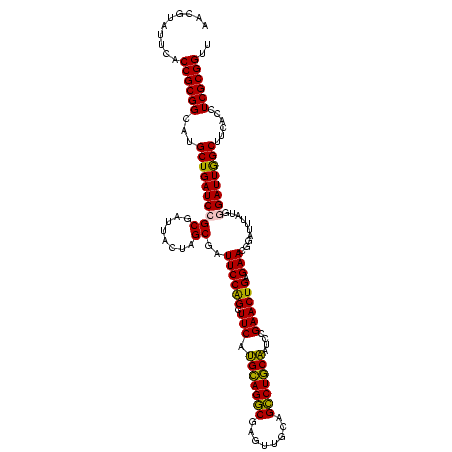
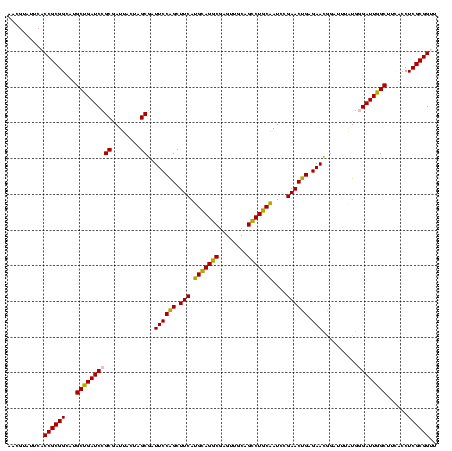
| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.42 |
| Mean single sequence MFE | -45.54 |
| Consensus MFE | -43.76 |
| Energy contribution | -44.16 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/1360-1480 CCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAA ..(((((..((....))..)))))((((.((.(((..((((((..((((...((.((((..(((..((.....)).))).)))).))..))))..))))))..)))))))))........ ( -45.60) >Bcere.0 330810 120 + 5224283/1360-1480 CCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAA ..(((((..((....))..)))))((((.((.(((..((((((..((((...((.((((..(((..((.....)).))).)))).))..))))..))))))..)))))))))........ ( -45.60) >Bhalo.0 1017954 120 + 4202352/1360-1480 CCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAA ..(((((..((....))..)))))((((.((.(((..((((((..((((...((.((((..(((..((.....)).))).)))).))..))))..))))))..)))))))))........ ( -45.60) >Bsubt.0 160926 120 + 4214630/1360-1480 CCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUUAGGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAA ..(((((..((....))..)))))((((.((.(((..((((((..((((.....((((((.(((..((.....)).)))...)))))).))))..))))))..)))))))))........ ( -45.30) >Blich.0 158327 119 + 4222334/1360-1480 CCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUU-GGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAA ..(((((..((....))..)))))((((.((.(((..((((((..((((.....((((((.(((..((....-)).)))...)))))).))))..))))))..)))))))))........ ( -45.60) >consensus CCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAA ..(((((..((....))..)))))((((.((.(((..((((((..((((...((.((((..(((..((.....)).))).)))).))..))))..))))))..)))))))))........ (-43.76 = -44.16 + 0.40)

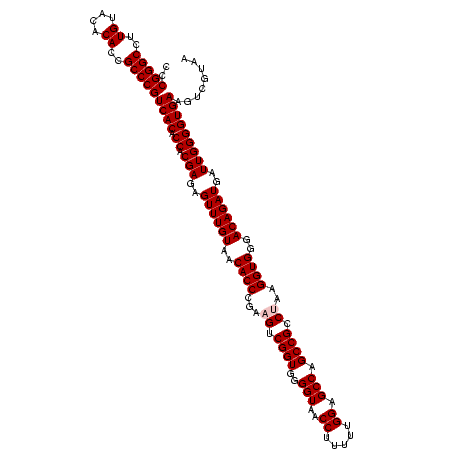
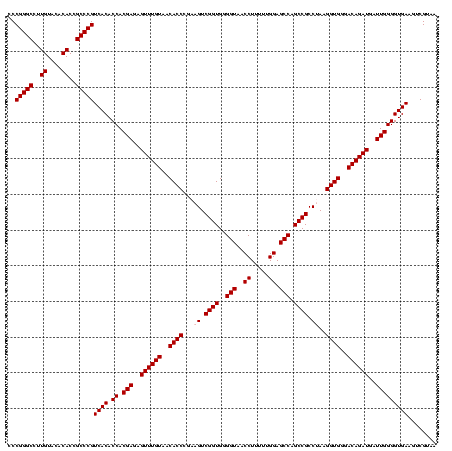
| Location | 290,091 – 290,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.42 |
| Mean single sequence MFE | -48.84 |
| Consensus MFE | -47.06 |
| Energy contribution | -47.46 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.87 |
| SVM RNA-class probability | 0.999675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 120 + 5227293/1400-1520 UGUAACACCCGAAGUCGGUGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCU (((..((((...((.((((..(((..((.....)).))).)))).))..))))..))).......(((((((.((....))....((((((((((....))))))))))..))))))).. ( -48.90) >Bcere.0 330810 120 + 5224283/1400-1520 UGUAACACCCGAAGUCGGUGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCU (((..((((...((.((((..(((..((.....)).))).)))).))..))))..))).......(((((((.((....))....((((((((((....))))))))))..))))))).. ( -48.90) >Bhalo.0 1017954 120 + 4202352/1400-1520 UGUAACACCCGAAGUCGGUGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCU (((..((((...((.((((..(((..((.....)).))).)))).))..))))..))).......(((((((.((....))....((((((((((....))))))))))..))))))).. ( -48.90) >Bsubt.0 160926 120 + 4214630/1400-1520 UGUAACACCCGAAGUCGGUGAGGUAACCUUUUAGGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCU (((..((((.....((((((.(((..((.....)).)))...)))))).))))..))).......(((((((.((....))....((((((((((....))))))))))..))))))).. ( -48.60) >Blich.0 158327 119 + 4222334/1400-1520 UGUAACACCCGAAGUCGGUGAGGUAACCUUUU-GGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCU (((..((((.....((((((.(((..((....-)).)))...)))))).))))..))).......(((((((.((....))....((((((((((....))))))))))..))))))).. ( -48.90) >consensus UGUAACACCCGAAGUCGGUGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCU (((..((((...((.((((..(((..((.....)).))).)))).))..))))..))).......(((((((.((....))....((((((((((....))))))))))..))))))).. (-47.06 = -47.46 + 0.40)

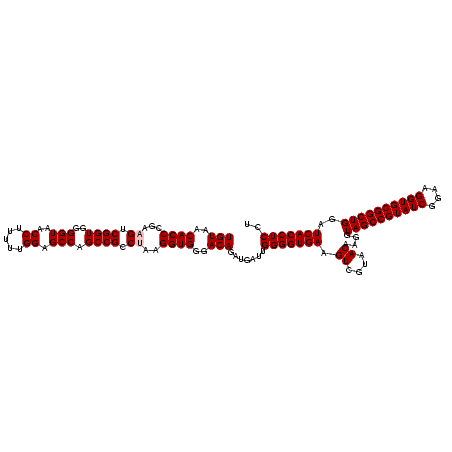
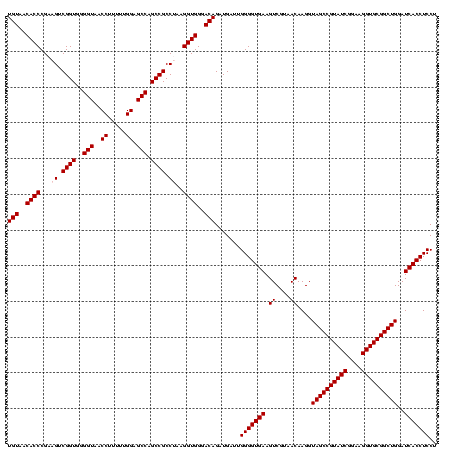
| Location | 290,091 – 290,210 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.14 |
| Mean single sequence MFE | -45.42 |
| Consensus MFE | -36.00 |
| Energy contribution | -36.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 119 + 5227293/1440-1560 GCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUU-GAUGAACGCUGUUCAUCAAUAUAAG .(((((...)))))..((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))....(((-(((((((...))))))))))..... ( -47.50) >Bcere.0 330810 119 + 5224283/1440-1560 GCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUU-GAUGAACGCUGUUCAUCAAUAUAAG .(((((...)))))..((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))....(((-(((((((...))))))))))..... ( -47.50) >Bhalo.0 1017954 120 + 4202352/1440-1560 GCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUACGGAGUUUUUAACUCUAGUCGACCAUUCGCAGAUU .(((((...)))))(((....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))...(((((.....))))).)))............... ( -43.60) >Bsubt.0 160926 118 + 4214630/1440-1560 GCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGA-UAUUAUACGGAAUAUAAGACCUUG-GGUCU .(((((...)))))(((........(((((((.((....))....((((((((((....))))))))))..)))))))....((((((.-..(((((.....)))))..)))))-)))). ( -45.00) >Blich.0 158327 119 + 4222334/1440-1560 GCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGA-UAUUAUACGGAAUAUAAGACCUUGCGGUCU .(((((...)))))(((....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))(((((.-..(((((.....)))))..)))))..))). ( -43.50) >consensus GCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAGUAUU_GACGAAAGAUGAGCAUUAGCAGAAU .(((((...))))).......((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..................................... (-36.00 = -36.00 + 0.00)

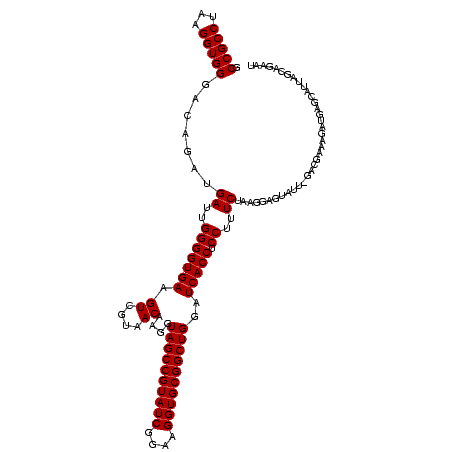
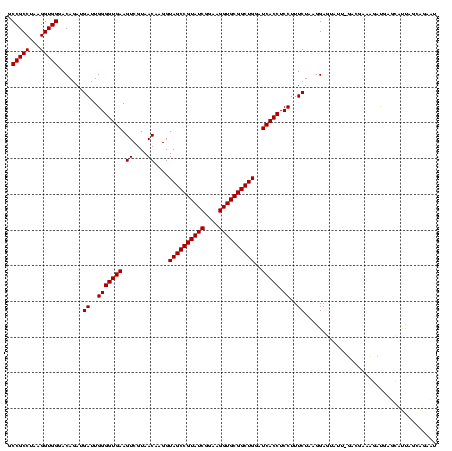
| Location | 290,091 – 290,199 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.29 |
| Mean single sequence MFE | -39.68 |
| Consensus MFE | -23.22 |
| Energy contribution | -23.38 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.59 |
| SVM decision value | 4.23 |
| SVM RNA-class probability | 0.999845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 108 + 5227293/1480-1600 CAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUU-GAUGAACGCUGUUCAUCAAUAUAAGUUUCCGUG--------UUUCG---UUUUGUUCAGUUUUGA (((..((((((((((....)))))))))(((.(((((((.......)))).(((-(((((((...))))))))))..........)))--------.))).---.........)..))). ( -38.30) >Bcere.0 330810 108 + 5224283/1480-1600 CAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUU-GAUGAACGCUGUUCAUCAAUAUAAGUUUCCGUG--------UUUCG---UUUUGUUCAGUUUUGA (((..((((((((((....)))))))))(((.(((((((.......)))).(((-(((((((...))))))))))..........)))--------.))).---.........)..))). ( -38.30) >Bhalo.0 1017954 120 + 4202352/1480-1600 CAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUACGGAGUUUUUAACUCUAGUCGACCAUUCGCAGAUUCAUCUGCGGAAGGACGCUUCGGACUUUUGUUCAGUUUUGA .((((.(((((((((....)))))))))((....)).)))).....(((((.....)))))((.((.((.((((((((....)))))))).)).))))..((((....))))........ ( -42.50) >Bsubt.0 160926 114 + 4214630/1480-1600 CAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGA-UAUUAUACGGAAUAUAAGACCUUG-GGUCUUAUAAACAG----AACGUUCCCUGUCUUGUUUAGUUUUGA ...((((((((((((....))))))))).....)))((((.....))))-.......((((((.(((((((...-)))))))((((((.----.(((.....)))..)))))))))))). ( -37.10) >Blich.0 158327 115 + 4222334/1480-1600 CAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGA-UAUUAUACGGAAUAUAAGACCUUGCGGUCUUAUAGACAG----GUGCGUUUGGAUCUUGUUUAGUUUUGA (((((((((((((((....)))))))))((((((((((((.....))))-........(...(((((((((....)))))))))..).)----))).))))..))))))........... ( -42.20) >consensus CAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAGUAUU_GACGAAAGAUGAGCAUUAGCAGAAUUAUCCGCGG_______CUUCG___UUUUGUUCAGUUUUGA (((((((((((((((....))))))))))......((((.......))))......................................................)))))........... (-23.22 = -23.38 + 0.16)

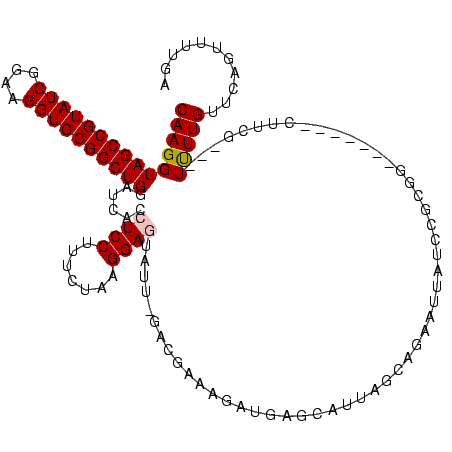
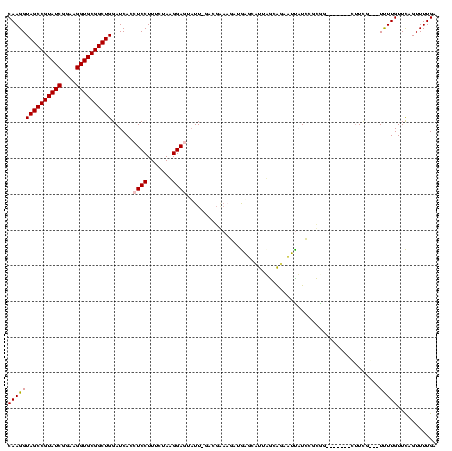
| Location | 290,091 – 290,199 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.29 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -14.40 |
| Energy contribution | -13.68 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 108 + 5227293/1480-1600 UCAAAACUGAACAAAA---CGAAA--------CACGGAAACUUAUAUUGAUGAACAGCGUUCAUC-AAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUG ................---.....--------...(((.......((((((((((...)))))))-)))(((((.......)))))...)))(((((............)))))...... ( -24.30) >Bcere.0 330810 108 + 5224283/1480-1600 UCAAAACUGAACAAAA---CGAAA--------CACGGAAACUUAUAUUGAUGAACAGCGUUCAUC-AAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUG ................---.....--------...(((.......((((((((((...)))))))-)))(((((.......)))))...)))(((((............)))))...... ( -24.30) >Bhalo.0 1017954 120 + 4202352/1480-1600 UCAAAACUGAACAAAAGUCCGAAGCGUCCUUCCGCAGAUGAAUCUGCGAAUGGUCGACUAGAGUUAAAAACUCCGUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUG ...............((((...((((.((...((((((....))))))...)).)).)).((((.....)))).(((....(((((((.........)))))))...)))))))...... ( -30.10) >Bsubt.0 160926 114 + 4214630/1480-1600 UCAAAACUAAACAAGACAGGGAACGUU----CUGUUUAUAAGACC-CAAGGUCUUAUAUUCCGUAUAAUA-UCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUG ................(((((......----.....(((((((((-...)))))))))..((((((....-((((.....))))((((.........)))).....))))))...))))) ( -28.60) >Blich.0 158327 115 + 4222334/1480-1600 UCAAAACUAAACAAGAUCCAAACGCAC----CUGUCUAUAAGACCGCAAGGUCUUAUAUUCCGUAUAAUA-UCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUG ...........((((.........(((----(....(((((((((....)))))))))............-((((.....))))))))....(((((............)))))..)))) ( -31.70) >consensus UCAAAACUGAACAAAA___CGAAA_______CCGCGGAUAAGUCUACAAAUGAUCAGCUUCCGUA_AAUACUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUG ....................................(.(((((((....))))))).).....................((((.((....))(((((............))))).)))). (-14.40 = -13.68 + -0.72)

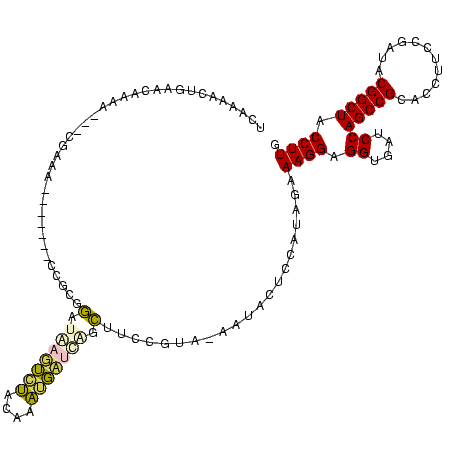
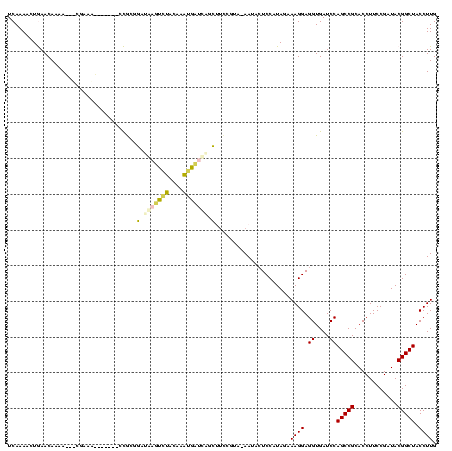
| Location | 290,091 – 290,196 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.80 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -14.02 |
| Energy contribution | -11.82 |
| Covariance contribution | -2.20 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 105 + 5227293/1497-1617 AAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUU-GAUGAACGCUGUUCAUCAAUAUAAGUUUCCGUG--------UUUCG---UUUUGUUCAGUUUUGAGAGAACU---AUCUCUC ((..((..((((((....((((.......)))).(((-(((((((...)))))))))).......)))).)--------)..))---..))..........((((((...---.)))))) ( -28.50) >Bcere.0 330810 105 + 5224283/1497-1617 AAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUU-GAUGAACGCUGUUCAUCAAUAUAAGUUUCCGUG--------UUUCG---UUUUGUUCAGUUUUGAGAGAACU---AUCUCUC ((..((..((((((....((((.......)))).(((-(((((((...)))))))))).......)))).)--------)..))---..))..........((((((...---.)))))) ( -28.50) >Bhalo.0 1017954 120 + 4202352/1497-1617 AAGGUGCGGCUGGAUCACCUCCUUUCUACGGAGUUUUUAACUCUAGUCGACCAUUCGCAGAUUCAUCUGCGGAAGGACGCUUCGGACUUUUGUUCAGUUUUGAGAGAACUCAAAACUCUC .(((((((((((((....((((.......))))........)))))))).((.((((((((....)))))))).)).))))).((((....))))(((((((((....)))))))))... ( -42.10) >Bsubt.0 160926 111 + 4214630/1497-1617 AAGGUGCGGCUGGAUCACCUCCUUUCUAAGGA-UAUUAUACGGAAUAUAAGACCUUG-GGUCUUAUAAACAG----AACGUUCCCUGUCUUGUUUAGUUUUGAAGGAUCA---UUCCUUC .(((.(((.(((.......((((.....))))-............(((((((((...-)))))))))..)))----..)))..)))...............((((((...---.)))))) ( -26.80) >Blich.0 158327 112 + 4222334/1497-1617 AAGGUGCGGCUGGAUCACCUCCUUUCUAAGGA-UAUUAUACGGAAUAUAAGACCUUGCGGUCUUAUAGACAG----GUGCGUUUGGAUCUUGUUUAGUUUUGAAGGAUCA---UUCCUUC (((((....(..(((((((((((.....))))-........(...(((((((((....)))))))))..).)----))).)))..))))))..........((((((...---.)))))) ( -31.20) >consensus AAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAGUAUU_GACGAAAGAUGAGCAUUAGCAGAAUUAUCCGCGG_______CUUCG___UUUUGUUCAGUUUUGAGAGAACU___AUCUCUC .......((((((((...((((.......))))...............(((.((....)).)))...........................))))))))..((((((.......)))))) (-14.02 = -11.82 + -2.20)

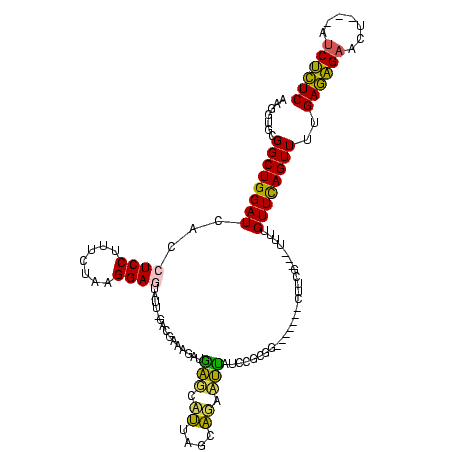
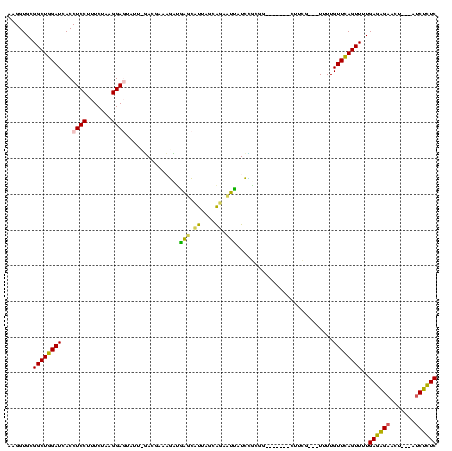
| Location | 290,091 – 290,196 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.80 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -17.12 |
| Energy contribution | -15.64 |
| Covariance contribution | -1.48 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 290091 105 + 5227293/1497-1617 GAGAGAU---AGUUCUCUCAAAACUGAACAAAA---CGAAA--------CACGGAAACUUAUAUUGAUGAACAGCGUUCAUC-AAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUU ((((((.---...))))))..............---.....--------...(((.......((((((((((...)))))))-)))(((((.......)))))...)))........... ( -26.80) >Bcere.0 330810 105 + 5224283/1497-1617 GAGAGAU---AGUUCUCUCAAAACUGAACAAAA---CGAAA--------CACGGAAACUUAUAUUGAUGAACAGCGUUCAUC-AAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUU ((((((.---...))))))..............---.....--------...(((.......((((((((((...)))))))-)))(((((.......)))))...)))........... ( -26.80) >Bhalo.0 1017954 120 + 4202352/1497-1617 GAGAGUUUUGAGUUCUCUCAAAACUGAACAAAAGUCCGAAGCGUCCUUCCGCAGAUGAAUCUGCGAAUGGUCGACUAGAGUUAAAAACUCCGUAGAAAGGAGGUGAUCCAGCCGCACCUU ...(((((((((....))))))))).........(((..((((.((...((((((....))))))...)).)).)).((((.....))))........)))((((.........)))).. ( -34.30) >Bsubt.0 160926 111 + 4214630/1497-1617 GAAGGAA---UGAUCCUUCAAAACUAAACAAGACAGGGAACGUU----CUGUUUAUAAGACC-CAAGGUCUUAUAUUCCGUAUAAUA-UCCUUAGAAAGGAGGUGAUCCAGCCGCACCUU ((((((.---...))))))....((((....(((((((....))----)))))(((((((((-...)))))))))............-...))))....((((((.........)))))) ( -28.50) >Blich.0 158327 112 + 4222334/1497-1617 GAAGGAA---UGAUCCUUCAAAACUAAACAAGAUCCAAACGCAC----CUGUCUAUAAGACCGCAAGGUCUUAUAUUCCGUAUAAUA-UCCUUAGAAAGGAGGUGAUCCAGCCGCACCUU ((((((.---...))))))......................(((----(....(((((((((....)))))))))............-((((.....))))))))............... ( -30.70) >consensus GAGAGAU___AGUUCUCUCAAAACUGAACAAAA___CGAAA_______CCGCGGAUAAGUCUACAAAUGAUCAGCUUCCGUA_AAUACUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUU ((((((.......))))))..................................(.(((((((....))))))).)........................((((((.........)))))) (-17.12 = -15.64 + -1.48)

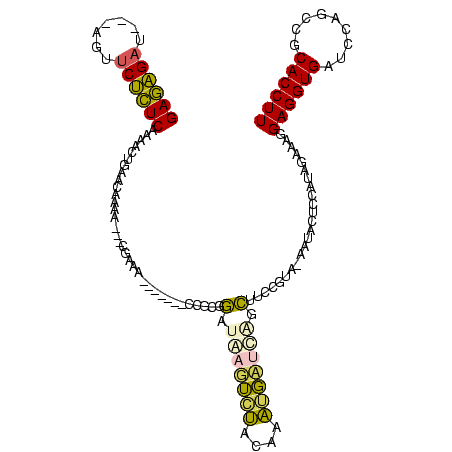
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:26:43 2006