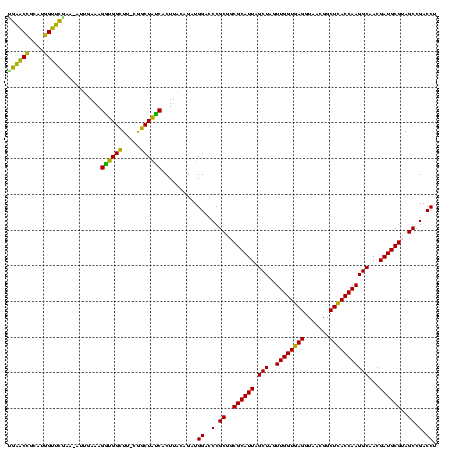
| Sequence ID | Bhalo.0 |
|---|---|
| Location | 656,180 – 656,300 |
| Length | 120 |
| Max. P | 0.999997 |

| Location | 656,180 – 656,298 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.12 |
| Mean single sequence MFE | -42.87 |
| Consensus MFE | -37.63 |
| Energy contribution | -35.50 |
| Covariance contribution | -2.13 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823201 |
| Prediction | RNA |
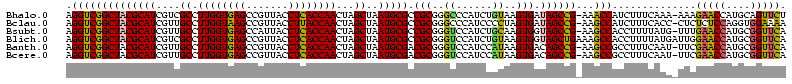
Download alignment: ClustalW | MAF
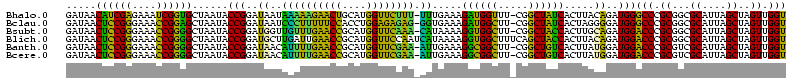
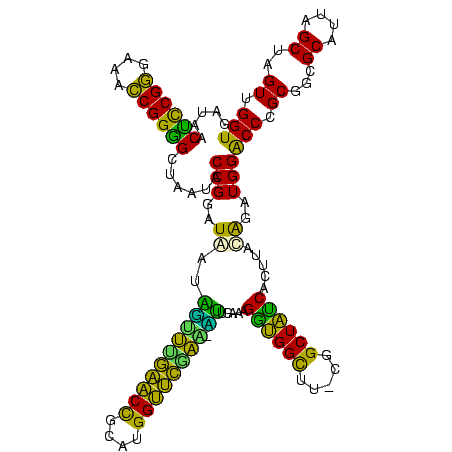
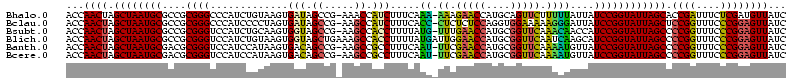
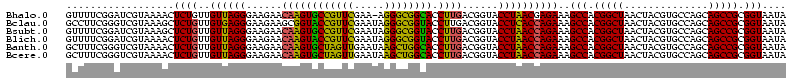
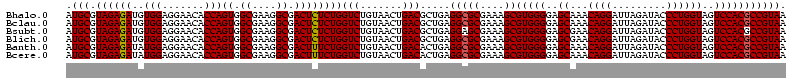
>Bhalo.0 656180 118 + 4202352/80-200 CGGACCAAA-GGGAGCUUGCUUCUG-GAGGUUAGCGGCGAACGGGUGAGUAACACGUGGGCAACCUGCCUGUAAGACUGGGAUAACAUCGAGAAAUCGGUGCUAAUACCGGAUAAUAAAA ..((((...-(((((....))))).-..)))).((((((.(((.((.....)).)))((....)))))).))....((((..((.((((((....)))))).))...))))......... ( -35.60) >Bclau.0 218057 118 + 4303871/80-200 CGGACAGAA-GGGAGCUUGCUCCCG-GACGUUAGCGGCGGACGGGUGAGUAACACGUGGGCAACCUGCCCCUUAGACUGGGAUAACUCCGGGAAACCGGAGCUAAUACCGGAUAAUCCCU .(((...((-(((.(((((((((((-..(((.....)))..))))..))))).....((....)).)))))))...((((..((.((((((....)))))).))...))))....))).. ( -50.80) >Bsubt.0 90519 118 + 4214630/80-200 CGGACAGAU-GGGAGCUUGCUCCCU-GAUGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGGUUGUU ..((((...-(((((....))))).-..))))((((((..((((((..((.((......)).))..))))))....((((..((.((((((....)))))).))...))))...)))))) ( -47.00) >Blich.0 95327 118 + 4222334/80-200 CGGACCGAC-GGGAGCUUGCUCUCU-UAGGUCAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGCUUGAU ..((((...-(((((....))))).-..))))((((.(..((((((..((.((......)).))..))))))..).((((..((.((((((....)))))).))...)))).)))).... ( -46.60) >Banth.0 82408 120 + 5227293/80-200 CGAAUGGAUUAAGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUU .(((((........(((..(((...((..(....)..))...)))..))).....((((((.....))))))....((((..((.((((((....)))))).))...))))....))))) ( -38.60) >Bcere.0 82410 120 + 5224283/80-200 CGAAUGGAUUAAGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUU .(((((........(((..(((...((..(....)..))...)))..))).....((((((.....))))))....((((..((.((((((....)))))).))...))))....))))) ( -38.60) >consensus CGGACAGAU_GGGAGCUUGCUCCCA_GAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAAUAGUU ..(((.....(((((....))))).....)))...((((.(((.((.....)).)))((....)))))).......((((..((.((((((....)))))).))...))))......... (-37.63 = -35.50 + -2.13)

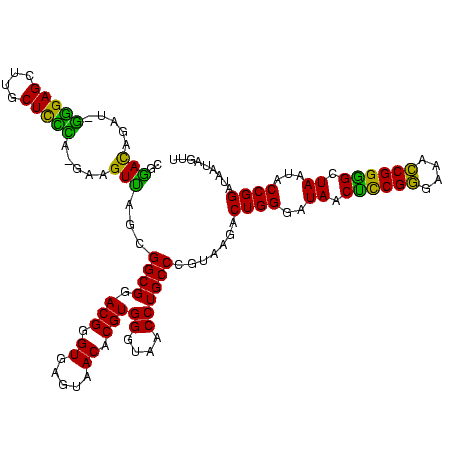

| Location | 656,180 – 656,298 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.78 |
| Mean single sequence MFE | -42.72 |
| Consensus MFE | -39.15 |
| Energy contribution | -35.38 |
| Covariance contribution | -3.77 |
| Combinations/Pair | 1.61 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 118 + 4202352/120-240 ACGGGUGAGUAACACGUGGGCAACCUGCCUGUAAGACUGGGAUAACAUCGAGAAAUCGGUGCUAAUACCGGAUAAUAAAAAGAACUGCAUGGUUCUUU-UUUGAAAGAUGGUUU-CGGCU (((.((.....)).)))((....)).(((.......((((..((.((((((....)))))).))...)))).....((((((((((....))))))))-)).((((.....)))-)))). ( -33.30) >Bclau.0 218057 118 + 4303871/120-240 ACGGGUGAGUAACACGUGGGCAACCUGCCCCUUAGACUGGGAUAACUCCGGGAAACCGGAGCUAAUACCGGAUAAUCCCUUUUUCCACCUGGAGAGAG-GGUGAAAGAUGGCUU-CGGCU (((.((.....)).)))((((.....))))(((...((((..((.((((((....)))))).))...)))).....((((((((((....))))))))-))...)))..(((..-..))) ( -49.20) >Bsubt.0 90519 118 + 4214630/120-240 ACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGCAUGGUUCAAA-CAUAAAAGGUGGCUU-CGGCU ((((((..((.((......)).))..))))))....((((..((.((((((....)))))).))...)))).....((((((((((....))))))))-))........(((..-..))) ( -46.30) >Blich.0 95327 120 + 4222334/120-240 ACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAACCGCAUGGUUCCAAUCAUAAAAGGUGGCUUUCAGCU ((((((..((.((......)).))..)))))).((.(((..(...((((((....))))))....((((.......((((((((((....)))).)))))).....))))..)..))))) ( -41.70) >Banth.0 82408 118 + 5227293/120-240 ACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAA-AUUGAAAGGCGGCUU-CGGCU ....(((.....)))((((((.....))))))....((((..((.((((((....)))))).))...)))).....((((((((((....))))))))-))........(((..-..))) ( -42.90) >Bcere.0 82410 118 + 5224283/120-240 ACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAA-AUUGAAAGGCGGCUU-CGGCU ....(((.....)))((((((.....))))))....((((..((.((((((....)))))).))...)))).....((((((((((....))))))))-))........(((..-..))) ( -42.90) >consensus ACGGGUGAGUAACACGUGGGUAACCUGCCCGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAAUAGUUUGAACCGCAUGGUUCGAA_AUUGAAAGGUGGCUU_CGGCU .((.(((.....))).))(((.....(((.......((((..((.((((((....)))))).))...)))).....((((((((((....)))))))).))........))).....))) (-39.15 = -35.38 + -3.77)
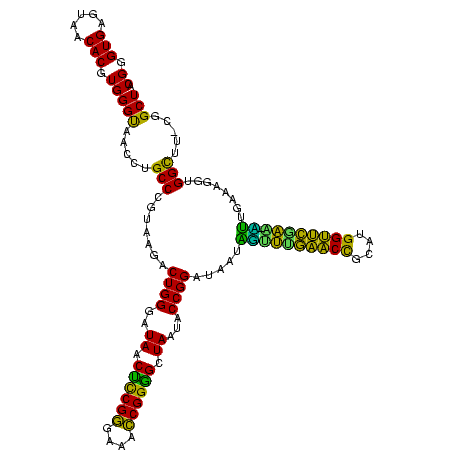
| Location | 656,180 – 656,298 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.98 |
| Mean single sequence MFE | -46.45 |
| Consensus MFE | -40.62 |
| Energy contribution | -35.83 |
| Covariance contribution | -4.79 |
| Combinations/Pair | 1.74 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 118 + 4202352/160-280 GAUAACAUCGAGAAAUCGGUGCUAAUACCGGAUAAUAAAAAGAACUGCAUGGUUCUUU-UUUGAAAGAUGGUUU-CGGCUAUCACUUACAGAUGGGCCCGCGGCGCAUUAGCUAGUUGGU ((((..(((((....)))))((..(.((((......((((((((((....))))))))-)).......)))).)-..))))))(((..(.((((.(((...))).)))).)..))).... ( -33.92) >Bclau.0 218057 118 + 4303871/160-280 GAUAACUCCGGGAAACCGGAGCUAAUACCGGAUAAUCCCUUUUUCCACCUGGAGAGAG-GGUGAAAGAUGGCUU-CGGCUAUCACUAGGGGAUGGGCCCGCGGCGCAUUAGCUAGUUGGU .....((((((....)))))).....((((......((((((((((....))))))))-)).....((((((..-..))))))(((((..((((.(((...))).))))..))))))))) ( -56.60) >Bsubt.0 90519 118 + 4214630/160-280 GAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGCAUGGUUCAAA-CAUAAAAGGUGGCUU-CGGCUACCACUUGCAGAUGGACCCGCGGCGCAUUAGCUAGUUGGU ..(((((((((....))))(((((((.(((...(((((((((((((....))))))))-)......((((((..-..))))))...........))))..)))...)))))))))))).. ( -51.70) >Blich.0 95327 120 + 4222334/160-280 GAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAACCGCAUGGUUCCAAUCAUAAAAGGUGGCUUUCAGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGU .....((((((....)))))).....(((((..(((....((..((((..(((.(((.((.(((..((((((.....))))))..)))..)))))))).))))..))..)))...))))) ( -45.50) >Banth.0 82408 118 + 5227293/160-280 GAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAA-AUUGAAAGGCGGCUU-CGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGU ..(((((((((....))))((((((((((((.....((((((((((....))))))))-)).....((((((..-..))))))..............))).))...)))))))))))).. ( -45.50) >Bcere.0 82410 118 + 5224283/160-280 GAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAA-AUUGAAAGGCGGCUU-CGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGU ..(((((((((....))))((((((((((((.....((((((((((....))))))))-)).....((((((..-..))))))..............))).))...)))))))))))).. ( -45.50) >consensus GAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAAUAGUUUGAACCGCAUGGUUCGAA_AUUGAAAGGUGGCUU_CGGCUAUCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGU .....((((((....))))))......(((..((..((((((((((....)))))))).)).....((((((.....)))))).....))..)))(((.((...((....))..)).))) (-40.62 = -35.83 + -4.79)
| Location | 656,180 – 656,298 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.98 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -25.59 |
| Energy contribution | -23.43 |
| Covariance contribution | -2.16 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.590898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 118 + 4202352/160-280 ACCAACUAGCUAAUGCGCCGCGGGCCCAUCUGUAAGUGAUAGCCG-AAACCAUCUUUCAAA-AAAGAACCAUGCAGUUCUUUUUAUUAUCCGGUAUUAGCACCGAUUUCUCGAUGUUAUC (((.(((.((..(((.(((...))).)))..)).)))(((((..(-(((.....)))).((-(((((((......))))))))).))))).)))..(((((.(((....))).))))).. ( -26.90) >Bclau.0 218057 118 + 4303871/160-280 ACCAACUAGCUAAUGCGCCGCGGGCCCAUCCCCUAGUGAUAGCCG-AAGCCAUCUUUCACC-CUCUCUCCAGGUGGAAAAAGGGAUUAUCCGGUAUUAGCUCCGGUUUCCCGGAGUUAUC (((.(((((...(((.(((...))).)))...)))))(((((..(-(((.....)))).((-((.(.(((....))).).)))).))))).)))..(((((((((....))))))))).. ( -37.10) >Bsubt.0 90519 118 + 4214630/160-280 ACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGCAAGUGGUAGCCG-AAGCCACCUUUUAUG-UUUGAACCAUGCGGUUCAAACAACCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUC ...((((.((((((((((..(((.(((((......)))).).)))-..)).........((-((((((((....))))))))))........)))))))).((((....))))))))... ( -42.10) >Blich.0 95327 120 + 4222334/160-280 ACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGGUAGCUGAAAGCCACCUUUUAUGAUUGGAACCAUGCGGUUCAAUCAAGCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUC ...((((.((((((((((((((((((((..((((((.(((.((.....)).))).))))))..))).))).)))))).........(....))))))))).((((....))))))))... ( -41.20) >Banth.0 82408 118 + 5227293/160-280 ACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCCG-AAGCCGCCUUUCAAU-UUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUC ...((((.(((((((((((....))).........((((((....-.((((((..(((...-...)))....)))))).....))))))...)))))))).((((....))))))))... ( -32.60) >Bcere.0 82410 118 + 5224283/160-280 ACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCCG-AAGCCGCCUUUCAAU-UUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUC ...((((.(((((((((((....))).........((((((....-.((((((..(((...-...)))....)))))).....))))))...)))))))).((((....))))))))... ( -32.60) >consensus ACCAACUAGCUAAUGCGCCGCGGGUCCAUCCGUAAGUGAUAGCCG_AAGCCACCUUUCAAG_UUCGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUC ...((((.((((((((....((((.............(((.((.....)).))).....((.((.(((((....))))).))))....)))))))))))).((((....))))))))... (-25.59 = -23.43 + -2.16)
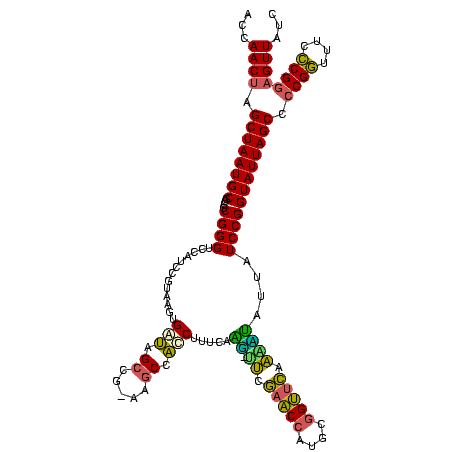
| Location | 656,180 – 656,298 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.97 |
| Mean single sequence MFE | -45.10 |
| Consensus MFE | -41.69 |
| Energy contribution | -39.28 |
| Covariance contribution | -2.41 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 118 + 4202352/200-320 AGAACUGCAUGGUUCUUU-UUUGAAAGAUGGUUU-CGGCUAUCACUUACAGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCU ((((((....))))))..-..........((((.-.(((((((.......))).)))).((.(((((((.(((..((((((((.......)))))))))))...))))))).)).)))). ( -40.10) >Bclau.0 218057 118 + 4303871/200-320 UUUUCCACCUGGAGAGAG-GGUGAAAGAUGGCUU-CGGCUAUCACUAGGGGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCAACGAUGCGUAGCCGACCU ((((((....)))))).(-(((....((((((..-..))))))(((((..((((.(((...))).))))..)))))(((((((.......))))))).(((.(((...))).))).)))) ( -46.80) >Bsubt.0 90519 118 + 4214630/200-320 UGAACCGCAUGGUUCAAA-CAUAAAAGGUGGCUU-CGGCUACCACUUGCAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCAACGAUGCGUAGCCGACCU ....((((..((((((..-..(((..((((((..-..))))))..)))....)))))).))))((((((.(((..((((((((.......)))))))))))...)))))).......... ( -45.70) >Blich.0 95327 120 + 4222334/200-320 UGAACCGCAUGGUUCCAAUCAUAAAAGGUGGCUUUCAGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCU ....((((..(((.(((.((.(((..((((((.....))))))..)))..)))))))).))))((((((.(((..((((((((.......)))))))))))...)))))).......... ( -43.60) >Banth.0 82408 118 + 5227293/200-320 UGAACCGCAUGGUUCGAA-AUUGAAAGGCGGCUU-CGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCU .(((((....)))))...-.......((((((..-..))))))......((.(((...(((((((.....(((..((((((((.......)))))))))))..)))))))...))).)). ( -47.20) >Bcere.0 82410 118 + 5224283/200-320 UGAACCGCAUGGUUCGAA-AUUGAAAGGCGGCUU-CGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCU .(((((....)))))...-.......((((((..-..))))))......((.(((...(((((((.....(((..((((((((.......)))))))))))..)))))))...))).)). ( -47.20) >consensus UGAACCGCAUGGUUCGAA_AUUGAAAGGUGGCUU_CGGCUAUCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCU ((((((....))))))..........((((((.....))))))..........((..(.((..((((((.(((..((((((((.......)))))))))))...))))))..)).).)). (-41.69 = -39.28 + -2.41)

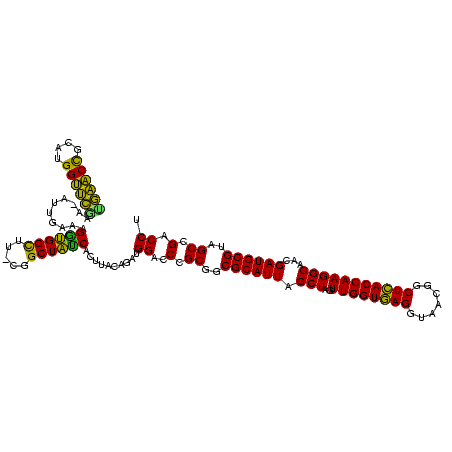
| Location | 656,180 – 656,298 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.97 |
| Mean single sequence MFE | -40.92 |
| Consensus MFE | -34.71 |
| Energy contribution | -33.93 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 118 + 4202352/200-320 AGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGCCCAUCUGUAAGUGAUAGCCG-AAACCAUCUUUCAAA-AAAGAACCAUGCAGUUCU .((((((((((((.........((((((((.......))))))))...((..(((.(((...))).)))..))..))).))))))-)..))..........-..(((((......))))) ( -35.40) >Bclau.0 218057 118 + 4303871/200-320 AGGUCGGCUACGCAUCGUUGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUGCGCCGCGGGCCCAUCCCCUAGUGAUAGCCG-AAGCCAUCUUUCACC-CUCUCUCCAGGUGGAAAA .((((((((((((...((.((.((((((((.......))))))))...((....)))).))(((......)))..))).))))))-..)))...(((((((-.........))))))).. ( -38.90) >Bsubt.0 90519 118 + 4214630/200-320 AGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCAUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGCAAGUGGUAGCCG-AAGCCACCUUUUAUG-UUUGAACCAUGCGGUUCA .((((((((((((((....((.((((((((.......))))))))...))..))))...(((((....))))).....)))))))-..)))..........-..((((((....)))))) ( -43.30) >Blich.0 95327 120 + 4222334/200-320 AGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGGUAGCUGAAAGCCACCUUUUAUGAUUGGAACCAUGCGGUUCA .((.((((........))))))((((((((.......))))))))...((....))((((((((((((..((((((.(((.((.....)).))).))))))..))).))).))))))... ( -44.10) >Banth.0 82408 118 + 5227293/200-320 AGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCCG-AAGCCGCCUUUCAAU-UUCGAACCAUGCGGUUCA (((.(((((.(((.((((.((.((((((((.......))))))))...))....)))).)))(((.(((......)))...))).-.))))))))......-...(((((....))))). ( -41.90) >Bcere.0 82410 118 + 5224283/200-320 AGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCCG-AAGCCGCCUUUCAAU-UUCGAACCAUGCGGUUCA (((.(((((.(((.((((.((.((((((((.......))))))))...))....)))).)))(((.(((......)))...))).-.))))))))......-...(((((....))))). ( -41.90) >consensus AGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCCGUAAGUGAUAGCCG_AAGCCACCUUUCAAG_UUCGAACCAUGCGGUUCA .((((((((((((((....((.((((((((.......))))))))...))..))))).(((..((......))..))).))))))...)))..............(((((....))))). (-34.71 = -33.93 + -0.77)
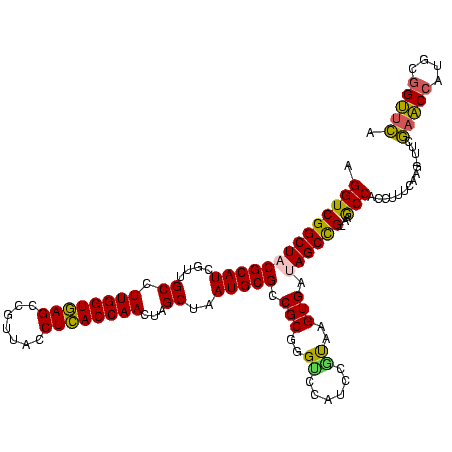
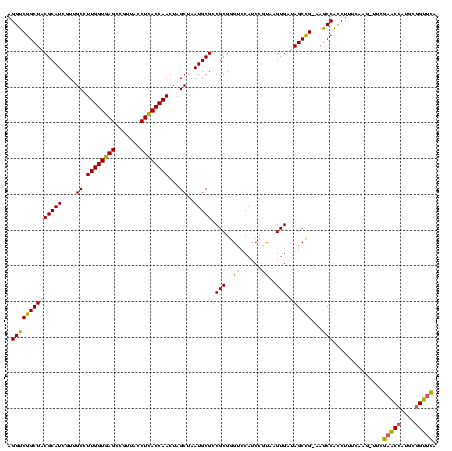
| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -48.75 |
| Consensus MFE | -46.65 |
| Energy contribution | -46.48 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.16 |
| SVM RNA-class probability | 0.999820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/240-360 UGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGCCCAUCUGUAAGUGAU ((((((..............((((((((((((.........))))))))))))..(((.((.((((((((.......))))))))...((....)))).)))))))))............ ( -50.00) >Bclau.0 218057 120 + 4303871/240-360 UGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUGCGCCGCGGGCCCAUCCCCUAGUGAU ((((((..............((((((((((((.........))))))))))))..(((.((.((((((((.......))))))))...((....)))).)))))))))............ ( -47.70) >Bsubt.0 90519 120 + 4214630/240-360 UGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCAUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGCAAGUGGU ((((((..............((((((((((((.........))))))))))))..(((.((.((((((((.......))))))))...((....)))).)))))))))............ ( -47.30) >Blich.0 95327 120 + 4222334/240-360 UGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGGU ((((((..............((((((((((((.........))))))))))))..(((.((.((((((((.......))))))))...((....)))).)))))))))............ ( -47.30) >Banth.0 82408 120 + 5227293/240-360 UGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGAC (((((((((((.((......((((((((((((.........))))))))))))...((((..((((((((.......)))))))).))))...)).))))).))))))............ ( -50.10) >Bcere.0 82410 120 + 5224283/240-360 UGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGAC (((((((((((.((......((((((((((((.........))))))))))))...((((..((((((((.......)))))))).))))...)).))))).))))))............ ( -50.10) >consensus UGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCCGUAAGUGAU ((((((..............((((((((((((.........))))))))))))..(((.((.((((((((.......))))))))...((....)))).)))))))))............ (-46.65 = -46.48 + -0.17)
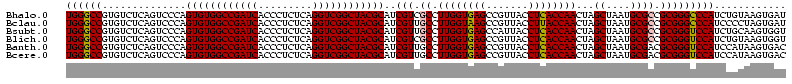
| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.89 |
| Mean single sequence MFE | -41.88 |
| Consensus MFE | -35.14 |
| Energy contribution | -38.00 |
| Covariance contribution | 2.86 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/360-480 GACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAG ..((((....))))..............((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....))))))..... ( -38.50) >Bclau.0 218057 120 + 4303871/360-480 GACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAGGAAGGCCUUCGGGUCGUAAAGCUCUGUUGUGAGGGAAGAAGCGG ..((((....))))...........(..((((((.((..((((....))))((((((((.....(((....))).....((((....)))).....)))))))).))..))))))..).. ( -50.20) >Bsubt.0 90519 120 + 4214630/360-480 GACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUUAGGGAAGAACAAG ..((((....))))..............((((((.....((((....))))((((((((..((.(((....))).))..((((....)))).....)))))))).....))))))..... ( -42.90) >Blich.0 95327 120 + 4222334/360-480 GACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAG ..((((....))))..............((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....))))))..... ( -38.50) >Banth.0 82408 120 + 5227293/360-480 GACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAG ..((((....))))..............((((((.....((((....))))(((((((..(((((.((.((((.......)))).)))).)))....))))))).....))))))..... ( -40.60) >Bcere.0 82410 120 + 5224283/360-480 GACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAG ..((((....))))..............((((((.....((((....))))(((((((..(((((.((.((((.......)))).)))).)))....))))))).....))))))..... ( -40.60) >consensus GACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAG ...((((((...........))))))..((((((.....((((....))))(((((((...(..(((....)))..)..((((....))))......))))))).....))))))..... (-35.14 = -38.00 + 2.86)

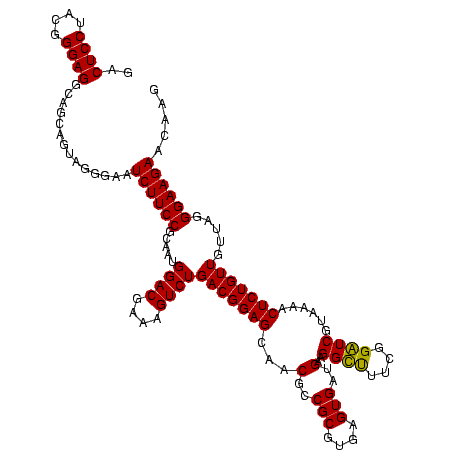
| Location | 656,180 – 656,299 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -47.52 |
| Consensus MFE | -43.53 |
| Energy contribution | -44.28 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.22 |
| SVM RNA-class probability | 0.999841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 119 + 4202352/400-520 GACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAA-AGGGCGGCACCUUGACGGUACCUAACGA (((....))).(((((((...((.(((....))).))..((((....))))......)))))))((((((......((((((((((((...-.)))))))).))))......)))))).. ( -48.80) >Bclau.0 218057 120 + 4303871/400-520 GACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAGGAAGGCCUUCGGGUCGUAAAGCUCUGUUGUGAGGGAAGAAGCGGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUCACCA (((....))).((((((((.....(((....))).....((((....)))).....))))))))((((((........((((((((((.....)))))))))).........)))))).. ( -57.53) >Bsubt.0 90519 120 + 4214630/400-520 GACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCA (((....))).((((((((..((.(((....))).))..((((....)))).....))))))))((((((......((((((((((((.....)))))))).))))......)))))).. ( -47.30) >Blich.0 95327 120 + 4222334/400-520 GACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCA (((....))).(((((((...((.(((....))).))..((((....))))......)))))))((((((......((((((((((((.....)))))))).))))......)))))).. ( -42.90) >Banth.0 82408 120 + 5227293/400-520 GACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCA (((....))).(((((((..(((((.((.((((.......)))).)))).)))....)))))))((((((......((((((((((((.....)))))))).))))......)))))).. ( -44.30) >Bcere.0 82410 120 + 5224283/400-520 GACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCA (((....))).(((((((..(((((.((.((((.......)))).)))).)))....)))))))((((((......((((((((((((.....)))))))).))))......)))))).. ( -44.30) >consensus GACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAAUAGGGCGGCACCUUGACGGUACCUAACCA (((....))).(((((((...(..(((....)))..)..((((....))))......)))))))((((((......((((((((((((.....)))))))).))))......)))))).. (-43.53 = -44.28 + 0.76)

| Location | 656,180 – 656,299 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -40.78 |
| Consensus MFE | -38.88 |
| Energy contribution | -36.94 |
| Covariance contribution | -1.94 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 119 + 4202352/440-560 GUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAA-AGGGCGGCACCUUGACGGUACCUAACGAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUA .((((((.(((((....(((((....))))).....((((((((((((...-.)))))))).)))))))))......)))))).(((.(((((..............))))).))).... ( -43.94) >Bclau.0 218057 120 + 4303871/440-560 GCCUUCGGGUCGUAAAGCUCUGUUGUGAGGGAAGAAGCGGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUCACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUA (((....)))........((((..((((((........((((((((((.....)))))))))).........))))))))))..(((.(((((..............))))).))).... ( -45.57) >Bsubt.0 90519 120 + 4214630/440-560 GUUUUCGGAUCGUAAAGCUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUA (((((((...)).)))))((((..((((((......((((((((((((.....)))))))).))))......))))))))))..(((.(((((..............))))).))).... ( -38.94) >Blich.0 95327 120 + 4222334/440-560 GUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUA (((((((...)).)))))((((..((((((......((((((((((((.....)))))))).))))......))))))))))..(((.(((((..............))))).))).... ( -38.94) >Banth.0 82408 120 + 5227293/440-560 GCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUA ((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..))))))(.(((((..............))))).)...... ( -38.64) >Bcere.0 82410 120 + 5224283/440-560 GCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUA ((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..))))))(.(((((..............))))).)...... ( -38.64) >consensus GCUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAAUAGGGCGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUA ..................((((..((((((......((((((((((((.....)))))))).))))......))))))))))..(((.(((((..............))))).))).... (-38.88 = -36.94 + -1.94)
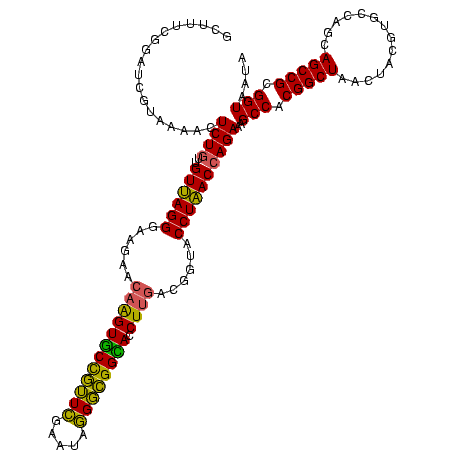
| Location | 656,180 – 656,299 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -38.59 |
| Consensus MFE | -35.69 |
| Energy contribution | -34.64 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 119 + 4202352/440-560 UAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUCGUUAGGUACCGUCAAGGUGCCGCCCU-UUCGAACGGCACUUGUUCUUCCCUAACAACAGAGUUUUACGAUCCGAAAAC .....((((((((((((.....))).))))))))).......((((((........((((((((..(.-...)..)))))))).......))))))......(((((.((...))))))) ( -36.96) >Bclau.0 218057 120 + 4303871/440-560 UAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUGAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGUACCGCUUCUUCCCUCACAACAGAGCUUUACGACCCGAAGGC .....((((((((((((.....))).)))))))))...((((((((((.........(((((((..(.....)..)))))))........))))))..))))(((((.((...))))))) ( -42.83) >Bsubt.0 90519 120 + 4214630/440-560 UAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGCACUUGUUCUUCCCUAACAACAGAGCUUUACGAUCCGAAAAC .....((((((((((((.....))).))))))))).((((.((((.((((((.....))))))(((..........)))....(((((...........))))).....)))).)))).. ( -36.90) >Blich.0 95327 120 + 4222334/440-560 UAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGUACUUGUUCUUCCCUAACAACAGAGUUUUACGAUCCGAAAAC .....((((((((((((.....))).)))))))))..(((((((((((........((((((((..(.....)..)))))))).......))))))..)))))................. ( -37.26) >Banth.0 82408 120 + 5227293/440-560 UAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCAGCUUAUUCAACUAGCACUUGUUCUUCCCUAACAACAGAGUUUUACGACCCGAAAGC ......(((((((((((.....))).))))))))((((((.((((.((((((.....)))))).(((.........)))..............................)))).)))))) ( -38.80) >Bcere.0 82410 120 + 5224283/440-560 UAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCAGCUUAUUCAACUAGCACUUGUUCUUCCCUAACAACAGAGUUUUACGACCCGAAAGC ......(((((((((((.....))).))))))))((((((.((((.((((((.....)))))).(((.........)))..............................)))).)))))) ( -38.80) >consensus UAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGCACUUGUUCUUCCCUAACAACAGAGUUUUACGACCCGAAAAC .....((((((((((((.....))).)))))))))...((((((((((........((((((((..(.....)..)))))))).......))))))..))))(((((.((...))))))) (-35.69 = -34.64 + -1.05)
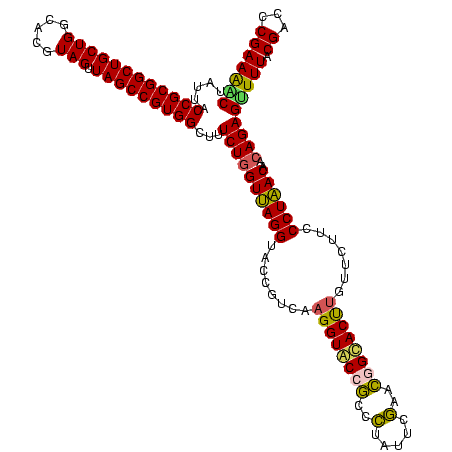
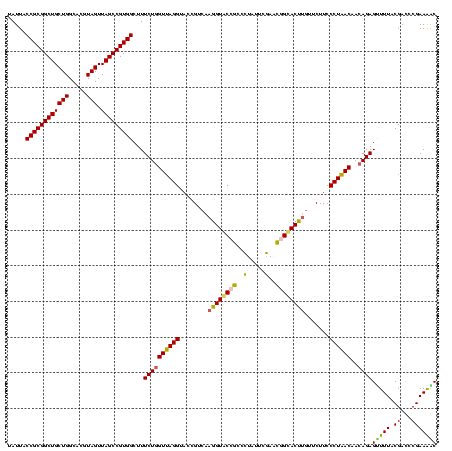
| Location | 656,180 – 656,299 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -39.72 |
| Consensus MFE | -37.51 |
| Energy contribution | -35.82 |
| Covariance contribution | -1.69 |
| Combinations/Pair | 1.15 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 119 + 4202352/480-600 UGCCGUUCGAA-AGGGCGGCACCUUGACGGUACCUAACGAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAUUAUUGGGCGUAAAG .(((((((...-.)))))))((((..(..((.((..(((((...(((((......(((((((((.((....)))))...)))))))))))...).))))..)).))..)..)).)).... ( -43.00) >Bclau.0 218057 120 + 4303871/480-600 UACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUCACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAUUAUUGGGCGUAAAG ...(((((((...((((((((((.....))))))..........((....((((.(((((((((.((....)))))...)))))).))))..))...)))).......)))))))..... ( -38.90) >Bsubt.0 90519 120 + 4214630/480-600 UGCCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAUUAUUGGGCGUAAAG .(((.((((.((((.((((((((.....))))))..........(((((......(((((((((.((....)))))...)))))))))))..)).)))).)))).......)))...... ( -40.10) >Blich.0 95327 120 + 4222334/480-600 UACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAUUAUUGGGCGUAAAG ...(((((((...((((((((((.....))))))..........((....((((.(((((((((.((....)))))...)))))).))))..))...)))).......)))))))..... ( -38.90) >Banth.0 82408 120 + 5227293/480-600 UGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUAUCCGGAAUUAUUGGGCGUAAAG (((...........((((((((......(((.....)))....(((....)))......)))))))).(((.(((((((.((..((((((....))))))))..)))))))))))))... ( -38.70) >Bcere.0 82410 120 + 5224283/480-600 UGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUAUCCGGAAUUAUUGGGCGUAAAG (((...........((((((((......(((.....)))....(((....)))......)))))))).(((.(((((((.((..((((((....))))))))..)))))))))))))... ( -38.70) >consensus UGCCGUUCGAAUAGGGCGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAUUAUUGGGCGUAAAG ((((((((.....)))))))).(((.(((...(((((((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).)).....)))))))).))) (-37.51 = -35.82 + -1.69)
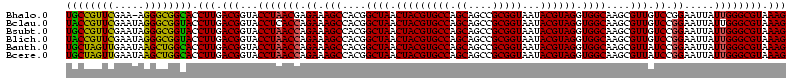

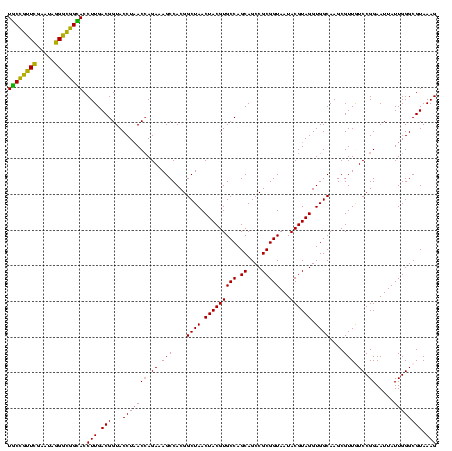
| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -42.20 |
| Consensus MFE | -41.22 |
| Energy contribution | -39.42 |
| Covariance contribution | -1.80 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/600-720 CGCGCGCAGGCGGUCUCUUAAGUCUGAUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGAGACUUGAGUACAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAA (((.((((.((..((((((...((((..((((....((((((.....))))))....)))).....(((((....))..))).))))..))))))((((....)))))).).)))))).. ( -44.90) >Bclau.0 218057 120 + 4303871/600-720 CGCGCGCAGGCGGCUUCUUAAGUCUGAUGUGAAAUCUCGGGGCUCAACCCCGAGCGGCCAUUGGAAACUGGGGAGCUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAA (((((((....(((((((((..((..(((.(....(((((((.....)))))))...))))..))...)))))))))...))))...........((((....)))).....)))..... ( -47.00) >Bsubt.0 90519 120 + 4214630/600-720 GGCUCGCAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAA .(((((((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))...........((((....))))..).)))...... ( -40.20) >Blich.0 95327 120 + 4222334/600-720 CGCGCGCAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAA (((((((....(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))...))))...........((((....)))).....)))..... ( -42.50) >Banth.0 82408 120 + 5227293/600-720 CGCGCGCAGGUGGUUUCUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAA (((((((....(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))...))))...........((((....)))).....)))..... ( -39.30) >Bcere.0 82410 120 + 5224283/600-720 CGCGCGCAGGUGGUUUCUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAA (((((((....(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))...))))...........((((....)))).....)))..... ( -39.30) >consensus CGCGCGCAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGAAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAA (((((((....(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))...))))...........((((....)))).....)))..... (-41.22 = -39.42 + -1.80)
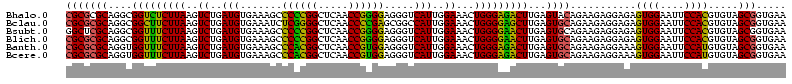
| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -27.20 |
| Energy contribution | -26.02 |
| Covariance contribution | -1.19 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/600-720 UUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGUACUCAAGUCUCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAGAGACCGCCUGCGCGCG .....(((.....((((....))))...........(((.....(((((..(((((....(((...(((((((.....)))))))...)))....)))))...)))))....)))))).. ( -34.00) >Bclau.0 218057 120 + 4303871/600-720 UUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGCUCCCCAGUUUCCAAUGGCCGCUCGGGGUUGAGCCCCGAGAUUUCACAUCAGACUUAAGAAGCCGCCUGCGCGCG .....(((.....((((....))))...........(((.....((((...(((((....(((...(((((((.....)))))))...)))....)))))...).)))....)))))).. ( -31.90) >Bsubt.0 90519 120 + 4214630/600-720 UUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUUCCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAGAAACCGCCUGCGAGCC ......(((....((((....))))..........((((......(((...(((((....(((...(((((((.....)))))))...)))....)))))...)))......))))))). ( -30.40) >Blich.0 95327 120 + 4222334/600-720 UUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUUCCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAGAAACCGCCUGCGCGCG .....(((.....((((....))))...........(((......(((...(((((....(((...(((((((.....)))))))...)))....)))))...)))......)))))).. ( -31.10) >Banth.0 82408 120 + 5227293/600-720 UUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAGAAACCACCUGCGCGCG .....(((.......((((.......))))......(((......(((...(((((....(((...(((((((.....)))))))...)))....)))))..))).......)))))).. ( -25.82) >Bcere.0 82410 120 + 5224283/600-720 UUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAGAAACCACCUGCGCGCG .....(((.......((((.......))))......(((......(((...(((((....(((...(((((((.....)))))))...)))....)))))..))).......)))))).. ( -25.82) >consensus UUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUCCCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAGAAACCGCCUGCGCGCG .....(((.....((((....))))...........(((.....(((.(..(((((....(((...(((((((.....)))))))...)))....)))))...).)))....)))..))) (-27.20 = -26.02 + -1.19)

| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -32.03 |
| Energy contribution | -31.62 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/640-760 GGCUCAACCGGGGAGGGUCAUUGGAAACUGGGAGACUUGAGUACAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGAC .((((((((.....))(((...(....).....)))))))))...........(.(((...((((((((.(.(((......)))....).))))))))...)))).((.((....)).)) ( -33.60) >Bclau.0 218057 120 + 4303871/640-760 GGCUCAACCCCGAGCGGCCAUUGGAAACUGGGGAGCUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGAC .((((((((((.((...((...))...)).))).).))))))...........(.(((...((((((((.(.(((......)))....).))))))))...)))).((.((....)).)) ( -37.70) >Bsubt.0 90519 120 + 4214630/640-760 GGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGAC .((((((((.....)).((...(....)....))..))))))...........(.(((...((((((((.(.(((......)))....).))))))))...)))).((.((....)).)) ( -32.40) >Blich.0 95327 120 + 4222334/640-760 GGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGAC .((((((((.....)).((...(....)....))..))))))...........(.(((...((((((((.(.(((......)))....).))))))))...)))).((.((....)).)) ( -32.40) >Banth.0 82408 120 + 5227293/640-760 GGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGAC .(((..(((((((((..((((((....).((....)).................)))))..))))))).)))))((((...(.((((....)))).)....)))).((.((....)).)) ( -33.40) >Bcere.0 82410 120 + 5224283/640-760 GGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGAC .(((..(((((((((..((((((....).((....)).................)))))..))))))).)))))((((...(.((((....)))).)....)))).((.((....)).)) ( -33.40) >consensus GGCUCAACCGGGGAGGGUCAUUGGAAACUGGGAAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGAC .(((...((.....))((((((((..(((((....))..))).............((....((((((((.(.(((......)))....).))))))))..)).))))))))...)))... (-32.03 = -31.62 + -0.41)

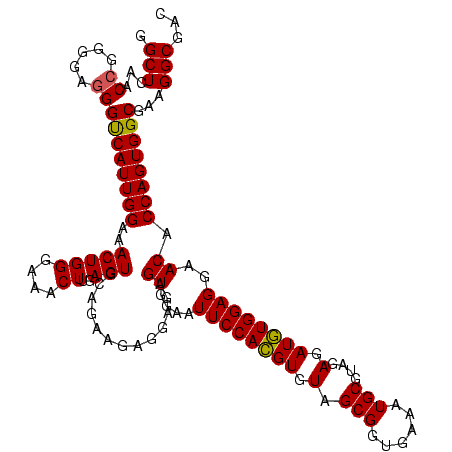
| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -44.90 |
| Consensus MFE | -44.64 |
| Energy contribution | -43.78 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.78 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.87 |
| SVM RNA-class probability | 0.999995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/680-800 GUACAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGG .((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..)))))).((.((((....))((....)))).)) ( -46.70) >Bclau.0 218057 120 + 4303871/680-800 GUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGG .((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..)))))).((.((((....))((....)))).)) ( -46.70) >Bsubt.0 90519 120 + 4214630/680-800 GUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGAGCGAAAGCGUGGG .((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..)))))).((.(((((..........))))).)) ( -45.50) >Blich.0 95327 120 + 4222334/680-800 GUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGG .((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..)))))).((.((((....))((....)))).)) ( -46.70) >Banth.0 82408 120 + 5227293/680-800 GUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGG .((((((..(((((.(((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))))............((((....))))... ( -41.90) >Bcere.0 82410 120 + 5224283/680-800 GUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGG .((((((..(((((.(((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))))............((((....))))... ( -41.90) >consensus GUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGG .((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))))............((((....))))... (-44.64 = -43.78 + -0.86)

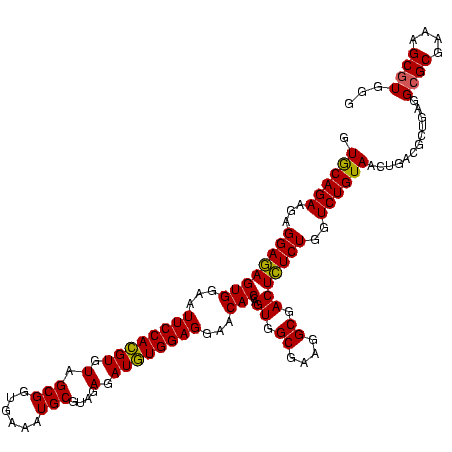

| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -24.44 |
| Energy contribution | -23.34 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.06 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/680-800 CCCACGCUUUCGCGCCUCAGCGUCAGUUACAGACCAGAGAGUCGCCUUCGCCACUGGUGUUCCUCCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGUAC .....(((..(((......)))..)))((((((...((((((.......((....(((((......))))).....))......((((.....))))......))))))....)))))). ( -29.10) >Bclau.0 218057 120 + 4303871/680-800 CCCACGCUUUCGCGCCUCAGCGUCAGUUACAGACCAGAGAGUCGCCUUCGCCACUGGUGUUCCUCCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCAC .....(((..(((......)))..)))..((((...((((((.......((....(((((......))))).....))......((((.....))))......))))))....))))... ( -26.30) >Bsubt.0 90519 120 + 4214630/680-800 CCCACGCUUUCGCUCCUCAGCGUCAGUUACAGACCAGAGAGUCGCCUUCGCCACUGGUGUUCCUCCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCAC .....(((..((((....))))..)))..((((...((((((.......((....(((((......))))).....))......((((.....))))......))))))....))))... ( -26.10) >Blich.0 95327 120 + 4222334/680-800 CCCACGCUUUCGCGCCUCAGCGUCAGUUACAGACCAGAGAGUCGCCUUCGCCACUGGUGUUCCUCCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCAC .....(((..(((......)))..)))..((((...((((((.......((....(((((......))))).....))......((((.....))))......))))))....))))... ( -26.30) >Banth.0 82408 120 + 5227293/680-800 CCCACGCUUUCGCGCCUCAGUGUCAGUUACAGACCAGAAAGUCGCCUUCGCCACUGGUGUUCCUCCAUAUCUCUACGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCAC ....(((....))).....(((.(((....(((...((((((((((.........))))....(((((........((........)).....))))).....)))))).))).)))))) ( -19.52) >Bcere.0 82410 120 + 5224283/680-800 CCCACGCUUUCGCGCCUCAGUGUCAGUUACAGACCAGAAAGUCGCCUUCGCCACUGGUGUUCCUCCAUAUCUCUACGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCAC ....(((....))).....(((.(((....(((...((((((((((.........))))....(((((........((........)).....))))).....)))))).))).)))))) ( -19.52) >consensus CCCACGCUUUCGCGCCUCAGCGUCAGUUACAGACCAGAGAGUCGCCUUCGCCACUGGUGUUCCUCCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCAC .....(((..(((......)))..)))..((((...((((((((((.........))))....(((((........((........)).....))))).....))))))....))))... (-24.44 = -23.34 + -1.11)

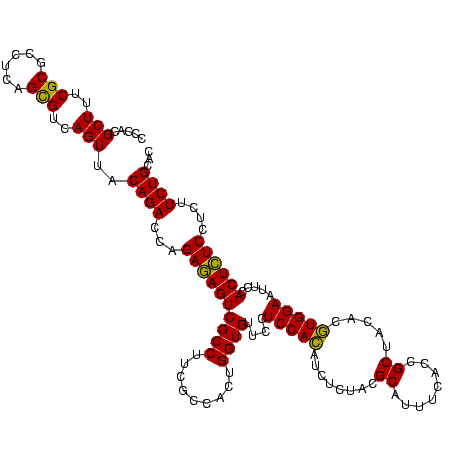
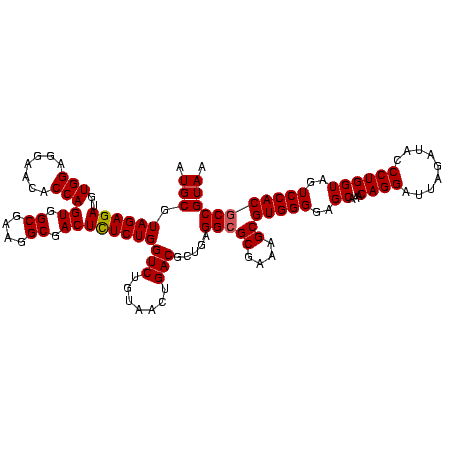
| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.94 |
| Mean single sequence MFE | -40.13 |
| Consensus MFE | -37.32 |
| Energy contribution | -37.27 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/720-840 AUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAA ..((.((((((.(((......)))..((.((....)).))))))))(((.......)))))...(((((....))(((((..((...((((.........))))))..)))))))).... ( -41.00) >Bclau.0 218057 120 + 4303871/720-840 AUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAA ..((.((((((.(((......)))..((.((....)).))))))))(((.......)))))...(((((....))(((((..((...((((.........))))))..)))))))).... ( -41.00) >Bsubt.0 90519 120 + 4214630/720-840 AUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGAGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAA ..(((((((((.(((......)))..((.((....)).)))))))((.((((..((.(((((..........))))).))..))...((((.........))))..)).))))))..... ( -37.90) >Blich.0 95327 120 + 4222334/720-840 AUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAA ..((.((((((.(((......)))..((.((....)).))))))))(((.......)))))...(((((....))((((...((...((((.........))))...))))))))).... ( -41.30) >Banth.0 82408 120 + 5227293/720-840 AUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAA ...........(((((.(((..(((((.(((....))((((....)))).(((.((((..(((..((((....)))).(....)...)))..)))).)))))))))..)))...))))). ( -39.80) >Bcere.0 82410 120 + 5224283/720-840 AUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAA ...........(((((.(((..(((((.(((....))((((....)))).(((.((((..(((..((((....)))).(....)...)))..)))).)))))))))..)))...))))). ( -39.80) >consensus AUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAA .(((.((((((..(((.......)))((.((....)).))))))))(((.......))).....(((((....))(((((..((...((((.........))))))..))))))))))). (-37.32 = -37.27 + -0.06)
| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -39.95 |
| Consensus MFE | -40.79 |
| Energy contribution | -39.57 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.70 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/760-880 UCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGACGCCCUUAGU ..............((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))((((......))))..... ( -39.40) >Bclau.0 218057 120 + 4303871/760-880 UCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGAUGCCCGUAGU ..............((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))((((......))))..... ( -40.60) >Bsubt.0 90519 120 + 4214630/760-880 UCUCUGGUCUGUAACUGACGCUGAGGAGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGU ..............((((((((....((((...((((((...((...((((.........))))...))))))))((....)).....)))).))))))))(((((....)))))..... ( -38.90) >Blich.0 95327 120 + 4222334/760-880 UCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGU ..............((((((((..(((((....((((((...((...((((.........))))...))))))))((....))....))))).))))))))(((((....)))))..... ( -40.20) >Banth.0 82408 120 + 5227293/760-880 UUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGU ..............((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))(((((....)))))..... ( -40.30) >Bcere.0 82410 120 + 5224283/760-880 UUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGU ..............((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))(((((....)))))..... ( -40.30) >consensus UCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCCUUAGU ..............((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))((((......))))..... (-40.79 = -39.57 + -1.22)


| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -40.95 |
| Consensus MFE | -41.98 |
| Energy contribution | -40.65 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.32 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/800-920 GAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGACGCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACG .......((((.........))))(((.(((..((((....))..(((((((.((..((((((((......))))))))..))...((....))....))))))).))....))).))). ( -40.50) >Bclau.0 218057 120 + 4303871/800-920 GAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGAUGCCCGUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACG .......((((.........))))(((.(((..((((....))..(((((((.((..(((.((((......)))).)))..))...((....))....))))))).))....))).))). ( -39.50) >Bsubt.0 90519 120 + 4214630/800-920 GAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACG .......((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))). ( -45.50) >Blich.0 95327 120 + 4222334/800-920 GAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGCAGCAAACGCAUUAAGCACUCCGCCUGGGGAGUACG .......((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))). ( -41.60) >Banth.0 82408 120 + 5227293/800-920 GAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACG .......((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))). ( -39.30) >Bcere.0 82410 120 + 5224283/800-920 GAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACG .......((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))). ( -39.30) >consensus GAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCCUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACG .......((((.........))))(((.(((..((((....))..(((((((.((..((((((((......))))))))..))...((....))....))))))).))....))).))). (-41.98 = -40.65 + -1.33)

| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.78 |
| Mean single sequence MFE | -50.52 |
| Consensus MFE | -52.27 |
| Energy contribution | -50.22 |
| Covariance contribution | -2.05 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.20 |
| Structure conservation index | 1.03 |
| SVM decision value | 6.10 |
| SVM RNA-class probability | 0.999997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/840-960 ACGAUGAGUGCUAGGUGUUAGGGGUUUCGACGCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGC .(((((((((((.((..((((((((......))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))). ( -49.10) >Bclau.0 218057 120 + 4303871/840-960 ACGAUGAGUGCUAGGUGUUAGGGGUUUCGAUGCCCGUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGACCCGC .(((((((((((.((..(((.((((......)))).)))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))). ( -50.10) >Bsubt.0 90519 120 + 4214630/840-960 ACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGC .(((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))). ( -54.00) >Blich.0 95327 120 + 4222334/840-960 ACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGCAGCAAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGC .(((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))). ( -50.10) >Banth.0 82408 120 + 5227293/840-960 ACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGC .(((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))). ( -49.90) >Bcere.0 82410 120 + 5224283/840-960 ACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGC .(((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))). ( -49.90) >consensus ACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCCUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGC .(((((((((((.((..((((((((......))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))). (-52.27 = -50.22 + -2.05)

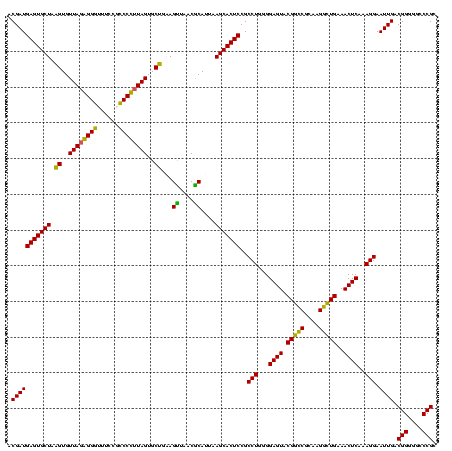
| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.78 |
| Mean single sequence MFE | -40.57 |
| Consensus MFE | -38.64 |
| Energy contribution | -38.03 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/880-1000 GCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCAGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGA ((((((.....(((((......((((((((...((((.(((((....)))))..))))..)))......)))))((((((.......)))).)))))))......))))..))....... ( -39.20) >Bclau.0 218057 120 + 4303871/880-1000 GCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCAGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGA ((((((...(((.(((...((.((((((((...((((.(((((....)))))..))))..)))......)))))..((((.......)))).))..))))))...))))..))....... ( -39.50) >Bsubt.0 90519 120 + 4214630/880-1000 GCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGA (((((.((....))(((((((.((((((((...((((.(((((....)))))..))))..)))......)))))((((((.......)))).)).....))))))).....)))..)).. ( -41.80) >Blich.0 95327 120 + 4222334/880-1000 GCUGCAGCAAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGA (((((.((....))(((((((.((((((((...((((.(((((....)))))..))))..)))......)))))((((((.......)))).)).....))))))).....)))..)).. ( -39.90) >Banth.0 82408 120 + 5227293/880-1000 GCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGA (((..(((((.(((((...((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))......)))))...)))))...)))....... ( -41.50) >Bcere.0 82410 120 + 5224283/880-1000 GCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGA (((..(((((.(((((...((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))......)))))...)))))...)))....... ( -41.50) >consensus GCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGA ((.(..((...((.(((((((.((((((((...((((.(((((....)))))..))))..)))......))))).....((((.((......)).))))))))))).))..))..))).. (-38.64 = -38.03 + -0.61)

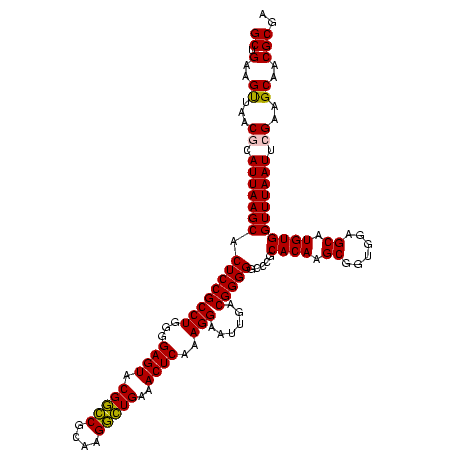
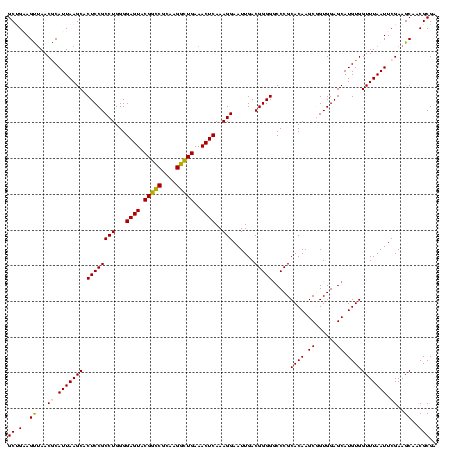
| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.79 |
| Mean single sequence MFE | -42.93 |
| Consensus MFE | -40.11 |
| Energy contribution | -41.67 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/1000-1120 AGAACCUUACCAGGUCUUGACAUCCUUUGACCACCCUAGAGAUAGGGCUUUCCCCUUCGGGGGACAAAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUA (((.(((....)))))).((((.((..(((((((((((....))).(((((((((....)))))..))))....)))))).)).)).))))...((((((..((....))..).))))). ( -44.50) >Bclau.0 218057 118 + 4303871/1000-1120 AGAACCUUACCAGGUCUUGACAUCCUUUGACCACCCAAGAGAUUGGGCUU-CCCCUUCGGGGG-CAAAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUA (((.(((....)))))).((((.((..(((((((((((....))).((((-((((....))))-..))))....)))))).)).)).))))...((((((..((....))..).))))). ( -42.20) >Bsubt.0 90519 118 + 4214630/1000-1120 AGAACCUUACCAGGUCUUGACAUCCUCUGACAAUCCUAGAGAUAGGACGU-CCCCUUCGGGGG-CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUA ...((((....))))((..((((((((((....(((((....)))))...-((((....))))-))))).((((.(.(.((.(((.....))))).).).))))....)))))..))... ( -42.50) >Blich.0 95327 118 + 4222334/1000-1120 AGAACCUUACCAGGUCUUGACAUCCUCUGACAACCCUAGAGAUAGGGCUU-CCCCUUCGGGGG-CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUA ..(((((...((..((.((((((...(((((..(((((....)))))((.-((((....))))-.))...(((((.((.....)).))))))))))...)))))).))..))..))))). ( -44.20) >Banth.0 82408 118 + 5227293/1000-1120 AGAACCUUACCAGGUCUUGACAUCCUCUGACAACCCUAGAGAUAGGGCUU-CUCCUUCGGGAG-CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUA ..(((((...((..((.((((((...(((((..(((((....)))))((.-((((....))))-.))...(((((.((.....)).))))))))))...)))))).))..))..))))). ( -42.10) >Bcere.0 82410 118 + 5224283/1000-1120 AGAACCUUACCAGGUCUUGACAUCCUCUGACAACCCUAGAGAUAGGGCUU-CUCCUUCGGGAG-CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUA ..(((((...((..((.((((((...(((((..(((((....)))))((.-((((....))))-.))...(((((.((.....)).))))))))))...)))))).))..))..))))). ( -42.10) >consensus AGAACCUUACCAGGUCUUGACAUCCUCUGACAACCCUAGAGAUAGGGCUU_CCCCUUCGGGGG_CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUA ...((((....))))((..((((((((((....(((((....)))))....((((....)))).))))).((((.(.(.((.(((.....))))).).).))))....)))))..))... (-40.11 = -41.67 + 1.56)
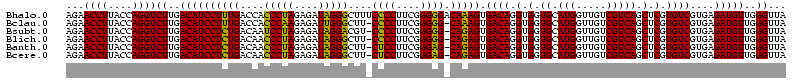
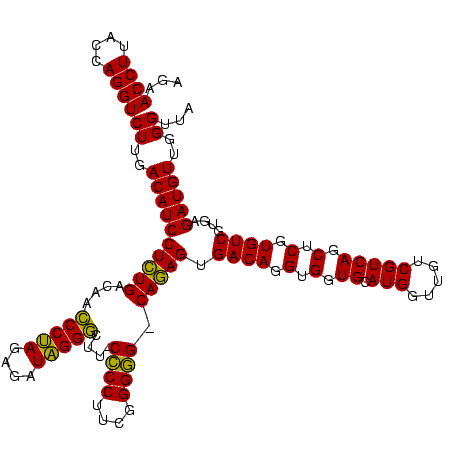

| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.79 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -31.24 |
| Energy contribution | -29.80 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/1000-1120 UAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGCACCACCUGUCACUUUGUCCCCCGAAGGGGAAAGCCCUAUCUCUAGGGUGGUCAAAGGAUGUCAAGACCUGGUAAGGUUCU .......(((((....((((.(.((((........)).))....).)))).(((((.(((((....)))...((((((....)))))))).))))))))))...(((((....))))).. ( -35.10) >Bclau.0 218057 118 + 4303871/1000-1120 UAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGCACCACCUGUCACUUUG-CCCCCGAAGGGG-AAGCCCAAUCUCUUGGGUGGUCAAAGGAUGUCAAGACCUGGUAAGGUUCU .......(((((....((((.(.((((........)).))....).)))).(((((-(((((....)))-..((((((....)))))))).))))))))))...(((((....))))).. ( -33.60) >Bsubt.0 90519 118 + 4214630/1000-1120 UAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGCACCACCUGUCACUCUG-CCCCCGAAGGGG-ACGUCCUAUCUCUAGGAUUGUCAGAGGAUGUCAAGACCUGGUAAGGUUCU .......((((((...((((.(((.(((((................))))))))..-((((....))))-..((((((....))))))))))...))))))...(((((....))))).. ( -31.29) >Blich.0 95327 118 + 4222334/1000-1120 UAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGCACCACCUGUCACUCUG-CCCCCGAAGGGG-AAGCCCUAUCUCUAGGGUUGUCAGAGGAUGUCAAGACCUGGUAAGGUUCU ................((((....(((((((((.............))))......-((((....))))-.(((((((....))))))))))))....))))..(((((....))))).. ( -34.22) >Banth.0 82408 118 + 5227293/1000-1120 UAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGCACCACCUGUCACUCUG-CUCCCGAAGGAG-AAGCCCUAUCUCUAGGGUUGUCAGAGGAUGUCAAGACCUGGUAAGGUUCU ................((((....(((((((((.............))))......-((((....))))-.(((((((....))))))))))))....))))..(((((....))))).. ( -32.12) >Bcere.0 82410 118 + 5224283/1000-1120 UAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGCACCACCUGUCACUCUG-CUCCCGAAGGAG-AAGCCCUAUCUCUAGGGUUGUCAGAGGAUGUCAAGACCUGGUAAGGUUCU ................((((....(((((((((.............))))......-((((....))))-.(((((((....))))))))))))....))))..(((((....))))).. ( -32.12) >consensus UAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGCACCACCUGUCACUCUG_CCCCCGAAGGGG_AAGCCCUAUCUCUAGGGUUGUCAGAGGAUGUCAAGACCUGGUAAGGUUCU .......(((((....((((.(.((((........)).))....).)))).(((((.((((....))))...((((((....))))))...))))))))))...(((((....))))).. (-31.24 = -29.80 + -1.44)

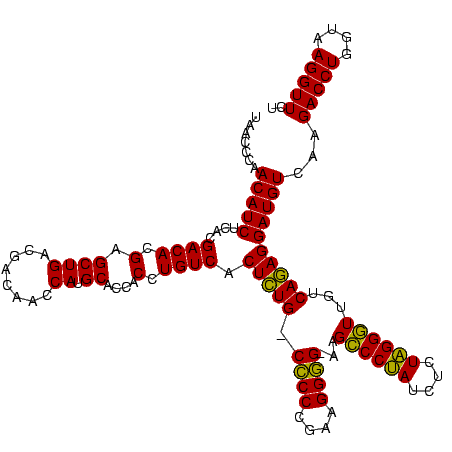

| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -36.07 |
| Consensus MFE | -35.31 |
| Energy contribution | -35.17 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.66 |
| Structure conservation index | 0.98 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/1080-1200 GGUUUGUCACCGGCAGUCACCUUAGAGUGCCCAACUGAAUGCUGGCAACUAAGGUCAAGGGUUGCGCUCGUUGCGGGACUUAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGC ((((.(((..((((.((((((((((..((((((......))..)))).)))))))...((((((.(((((...)))).).))))))..........)))....))))..))))))).... ( -39.30) >Bclau.0 218057 120 + 4303871/1080-1200 GGUUUGUCACCGGCAGUCACCUUAGAGUGCCCAACUCAAUGCUGGCAACUAAGAUCAAGGGUUGCGCUCGUUGCGGGACUUAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGC ((((.(((...((((.((......)).))))....(((......((((((.........))))))((((((...(((......)))....((....)).))))))))).))))))).... ( -36.30) >Bsubt.0 90519 120 + 4214630/1080-1200 GGUUUGUCACCGGCAGUCACCUUAGAGUGCCCAACUGAAUGCUGGCAACUAAGAUCAAGGGUUGCGCUCGUUGCGGGACUUAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGC ((((.(((..((((.(((......((((((((...(((.....(....).....)))..))..))))))((((.((.......)))))).......)))....))))..))))))).... ( -35.60) >Blich.0 95327 120 + 4222334/1080-1200 GGUUUGUCACCGGCAGUCACCUUAGAGUGCCCAACUGAAUGCUGGCAACUAAGAUCAAGGGUUGCGCUCGUUGCGGGACUUAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGC ((((.(((..((((.(((......((((((((...(((.....(....).....)))..))..))))))((((.((.......)))))).......)))....))))..))))))).... ( -35.60) >Banth.0 82408 120 + 5227293/1080-1200 GGUUUGUCACCGGCAGUCACCUUAGAGUGCCCAACUUAAUGAUGGUAACUAAGAUCAAGGGUUGCGCUCGUUGCGGGACUUAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGC ((((.(((..((((.(((......((((((..(((((..((((..........)))).)))))))))))((((.((.......)))))).......)))....))))..))))))).... ( -34.00) >Bcere.0 82410 120 + 5224283/1080-1200 GGUUUGUCACCGGCAGUCACCUUAGAGUGCCCAACUGAAUGAUGGCAACUAAGAUCAAGGGUUGCGCUCGUUGCGGGACUUAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGC ((((.(((..((((.(((......((((((((...(((.....(....).....)))..))..))))))((((.((.......)))))).......)))....))))..))))))).... ( -35.60) >consensus GGUUUGUCACCGGCAGUCACCUUAGAGUGCCCAACUGAAUGCUGGCAACUAAGAUCAAGGGUUGCGCUCGUUGCGGGACUUAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGC ((((.(((...((((.((......)).)))).............((((((.........))))))((((((...(((......)))....((....)).))))))....))))))).... (-35.31 = -35.17 + -0.14)

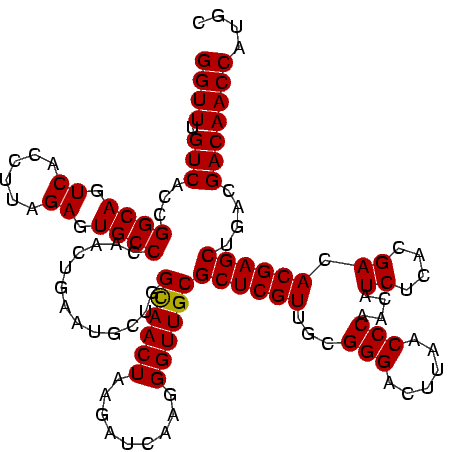
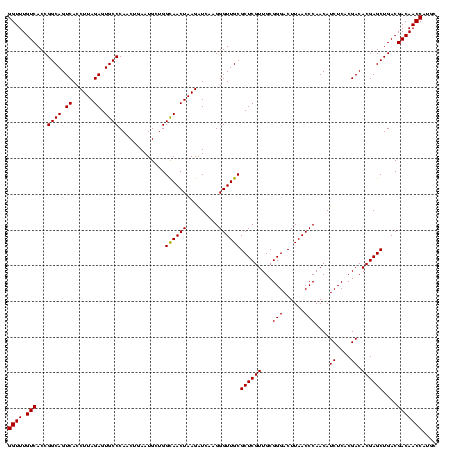
| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -39.75 |
| Consensus MFE | -38.62 |
| Energy contribution | -38.98 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/1120-1240 AGUCCCGCAACGAGCGCAACCCUUGACCUUAGUUGCCAGCAUUCAGUUGGGCACUCUAAGGUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUA .((..(((.....)))..))((((.(((((((.(((((((.....))).))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).))))... ( -43.10) >Bclau.0 218057 120 + 4303871/1120-1240 AGUCCCGCAACGAGCGCAACCCUUGAUCUUAGUUGCCAGCAUUGAGUUGGGCACUCUAAGGUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUA .((..(((.....)))..))((((.(((((((.(((((((.....))).))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).))))... ( -40.50) >Bsubt.0 90519 120 + 4214630/1120-1240 AGUCCCGCAACGAGCGCAACCCUUGAUCUUAGUUGCCAGCAUUCAGUUGGGCACUCUAAGGUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUA .((..(((.....)))..))((((.(((((((.(((((((.....))).))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).))))... ( -40.50) >Blich.0 95327 120 + 4222334/1120-1240 AGUCCCGCAACGAGCGCAACCCUUGAUCUUAGUUGCCAGCAUUCAGUUGGGCACUCUAAGGUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUA .((..(((.....)))..))((((.(((((((.(((((((.....))).))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).))))... ( -40.50) >Banth.0 82408 120 + 5227293/1120-1240 AGUCCCGCAACGAGCGCAACCCUUGAUCUUAGUUACCAUCAUUAAGUUGGGCACUCUAAGGUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUA .((..(((.....)))..))((((.(((((((...(((.(.....).))).....)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).))))... ( -35.30) >Bcere.0 82410 120 + 5224283/1120-1240 AGUCCCGCAACGAGCGCAACCCUUGAUCUUAGUUGCCAUCAUUCAGUUGGGCACUCUAAGGUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUA .((..(((.....)))..))((((.(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).))))... ( -38.60) >consensus AGUCCCGCAACGAGCGCAACCCUUGAUCUUAGUUGCCAGCAUUCAGUUGGGCACUCUAAGGUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUA .((..(((.....)))..))((((.(((((((.(((((((.....))).))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).))))... (-38.62 = -38.98 + 0.36)

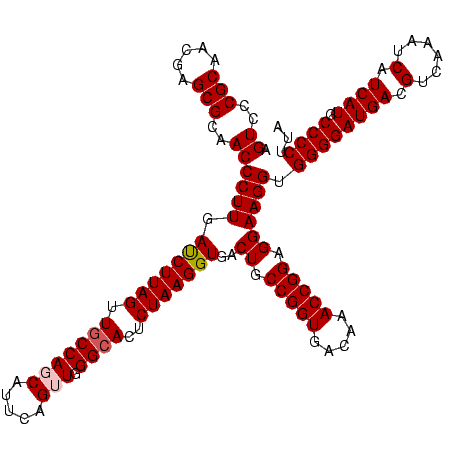
| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -41.83 |
| Consensus MFE | -41.34 |
| Energy contribution | -41.20 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/1120-1240 UAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCACCUUAGAGUGCCCAACUGAAUGCUGGCAACUAAGGUCAAGGGUUGCGCUCGUUGCGGGACU ...((((.((((((......)))))))))).((((.(((((((.....))))).))..(((((((..((((((......))..)))).))))))).))))(((.(((.....))).))). ( -45.50) >Bclau.0 218057 120 + 4303871/1120-1240 UAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCACCUUAGAGUGCCCAACUCAAUGCUGGCAACUAAGAUCAAGGGUUGCGCUCGUUGCGGGACU ...((((.((((((......)))))))))).((((.(((((((.....))))).))....(((((..((((((......))..)))).)))))...))))(((.(((.....))).))). ( -41.50) >Bsubt.0 90519 120 + 4214630/1120-1240 UAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCACCUUAGAGUGCCCAACUGAAUGCUGGCAACUAAGAUCAAGGGUUGCGCUCGUUGCGGGACU ...((((.((((((......)))))))))).((((.(((((((.....))))).))....(((((..((((((......))..)))).)))))...))))(((.(((.....))).))). ( -41.50) >Blich.0 95327 120 + 4222334/1120-1240 UAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCACCUUAGAGUGCCCAACUGAAUGCUGGCAACUAAGAUCAAGGGUUGCGCUCGUUGCGGGACU ...((((.((((((......)))))))))).((((.(((((((.....))))).))....(((((..((((((......))..)))).)))))...))))(((.(((.....))).))). ( -41.50) >Banth.0 82408 120 + 5227293/1120-1240 UAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCACCUUAGAGUGCCCAACUUAAUGAUGGUAACUAAGAUCAAGGGUUGCGCUCGUUGCGGGACU ...((((.((((((......)))))))))).((((.(((((((.....))))).))....(((((..((((((......))..)))).)))))...))))(((.(((.....))).))). ( -39.50) >Bcere.0 82410 120 + 5224283/1120-1240 UAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCACCUUAGAGUGCCCAACUGAAUGAUGGCAACUAAGAUCAAGGGUUGCGCUCGUUGCGGGACU ...((((.((((((......)))))))))).((((.(((((((.....))))).))....(((((..((((((......))..)))).)))))...))))(((.(((.....))).))). ( -41.50) >consensus UAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCACCUUAGAGUGCCCAACUGAAUGCUGGCAACUAAGAUCAAGGGUUGCGCUCGUUGCGGGACU ...((((.((((((......)))))))))).((((.(((((((.....))))).))....(((((..((((((......))..)))).)))))...))))(((.(((.....))).))). (-41.34 = -41.20 + -0.14)

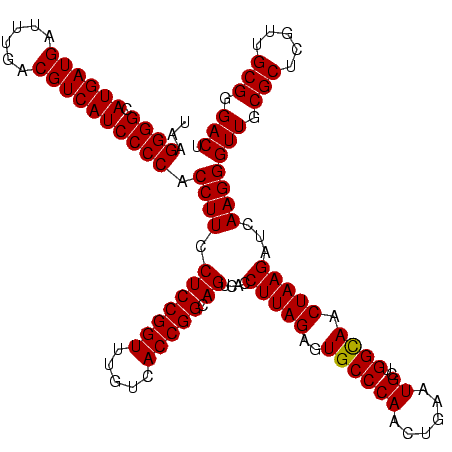
| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -41.61 |
| Energy contribution | -37.90 |
| Covariance contribution | -3.71 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.35 |
| Structure conservation index | 1.08 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/1240-1360 UGACCUGGGCUACACACGUGCUACAAUGGAUGGUACAAAGGGUUGCGAAGCCGCGAGGUGAAGCCAAUCCCAGAAAGCCAUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAA .((.((((((.((....))))).....(((((((.....(((((((...(((....)))...).))))))......))))))).))).))....(((((((........))))))).... ( -41.40) >Bclau.0 218057 120 + 4303871/1240-1360 UGACCUGGGCUACACACGUGCUACAAUGGAUGGUACAAAGGGCAGCGAAACCGCGAGGUGAAGCCAAUCCCAUAAAGCCAUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAA .((.((((((.((....))))).....(((((((.....(((..((...(((....)))...))....))).....))))))).))).))....(((((((........))))))).... ( -39.90) >Bsubt.0 90519 120 + 4214630/1240-1360 UGACCUGGGCUACACACGUGCUACAAUGGACAGAACAAAGGGCAGCGAAACCGCGAGGUUAAGCCAAUCCCACAAAUCUGUUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAA .((.(((((((.((....)).......(((((((.....(((..((..((((....))))..))....))).....)))))))..))))))).))((((((........))))))..... ( -39.00) >Blich.0 95327 120 + 4222334/1240-1360 UGACCUGGGCUACACACGUGCUACAAUGGGCAGAACAAAGGGCAGCGAAGCCGCGAGGCUAAGCCAAUCCCACAAAUCUGUUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAA .((.(((((((.((....)).......(((((((.....(((..((..((((....))))..))....))).....)))))))..))))))).))((((((........))))))..... ( -40.90) >Banth.0 82408 120 + 5227293/1240-1360 UGACCUGGGCUACACACGUGCUACAAUGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAA .((.((((((.((....))))).....(((((((.....(((..((...(((....)))...))....))).....))))))).))).))....(((((((........))))))).... ( -35.40) >Bcere.0 82410 120 + 5224283/1240-1360 UGACCUGGGCUACACACGUGCUACAAUGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAA .((.((((((.((....))))).....(((((((.....(((..((...(((....)))...))....))).....))))))).))).))....(((((((........))))))).... ( -35.40) >consensus UGACCUGGGCUACACACGUGCUACAAUGGACGGUACAAAGGGCAGCGAAACCGCGAGGUGAAGCCAAUCCCAUAAAACCGUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAA .((.((((((.((....))))).....(((((((.....(((..((...(((....)))...))....))).....))))))).))).))....(((((((........))))))).... (-41.61 = -37.90 + -3.71)


| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.67 |
| Mean single sequence MFE | -42.60 |
| Consensus MFE | -39.34 |
| Energy contribution | -37.90 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/1280-1400 CACCGCGGCAUGCUGAUCCGCGAUUACUAGCAAUUCCGGCUUCAUGCAGGCGAGUUGCAGCCUGCAAUCCGAACUGAGAAUGGCUUUCUGGGAUUGGCUUCACCUCGCGGCUUCGCAACC ..((((((...((..((((..((.....(((.(((((((.(((.(((((((........)))))))....)))))).)))).))).))..))))..))......)))))).......... ( -42.90) >Bclau.0 218057 120 + 4303871/1280-1400 CACCGCGGCAUGCUGAUCCGCGAUUACUAGCAAUUCCGGCUUCAUGCAGGCGAGUUGCAGCCUGCAAUCCGAACUGAGAAUGGCUUUAUGGGAUUGGCUUCACCUCGCGGUUUCGCUGCC ......((((.((.((.((((((.....(((((((((((((((.(((((((........))))))).((......))))..))))....)))))).))).....)))))).)).)))))) ( -44.00) >Bsubt.0 90519 120 + 4214630/1280-1400 CACCGCGGCAUGCUGAUCCGCGAUUACUAGCGAUUCCAGCUUCACGCAGUCGAGUUGCAGACUGCGAUCCGAACUGAGAACAGAUUUGUGGGAUUGGCUUAACCUCGCGGUUUCGCUGCC ......((((.((.((.((((((.....((((((((((......(((((((........)))))))...(((((((....))).))))))))))).))).....)))))).)).)))))) ( -45.70) >Blich.0 95327 120 + 4222334/1280-1400 CACCGCGGCAUGCUGAUCCGCGAUUACUAGCGAUUCCAGCUUCACGCAGUCGAGUUGCAGACUGCGAUCCGAACUGAGAACAGAUUUGUGGGAUUGGCUUAGCCUCGCGGCUUCGCUGCC ......((((.((.((.((((((.....((((((((((......(((((((........)))))))...(((((((....))).))))))))))).))).....)))))).)).)))))) ( -45.60) >Banth.0 82408 120 + 5227293/1280-1400 CACCGCGGCAUGCUGAUCCGCGAUUACUAGCGAUUCCAGCUUCAUGUAGGCGAGUUGCAGCCUACAAUCCGAACUGAGAACGGUUUUAUGAGAUUAGCUCCACCUCGCGGUCUUGCAGCU .(((((((...((((((((((........))).((((((.(((.(((((((........)))))))....)))))).)))...........)))))))......)))))))......... ( -38.70) >Bcere.0 82410 120 + 5224283/1280-1400 CACCGCGGCAUGCUGAUCCGCGAUUACUAGCGAUUCCAGCUUCAUGUAGGCGAGUUGCAGCCUACAAUCCGAACUGAGAACGGUUUUAUGAGAUUAGCUCCACCUCGCGGUCUUGCAGCU .(((((((...((((((((((........))).((((((.(((.(((((((........)))))))....)))))).)))...........)))))))......)))))))......... ( -38.70) >consensus CACCGCGGCAUGCUGAUCCGCGAUUACUAGCGAUUCCAGCUUCAUGCAGGCGAGUUGCAGCCUGCAAUCCGAACUGAGAACGGAUUUAUGGGAUUGGCUUCACCUCGCGGUUUCGCAGCC ..((((((...((((((((((........))..((((((.(((.(((((((........)))))))....)))))).)))..........))))))))......)))))).......... (-39.34 = -37.90 + -1.44)
| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -40.42 |
| Consensus MFE | -41.07 |
| Energy contribution | -39.38 |
| Covariance contribution | -1.69 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.06 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/1320-1440 UUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAAGCCGGAAUUGCUAGUAAUCGCGGAUCAGCAUGCCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCA ....((((((((..(((((((........))))))).....)).))))))...((((((((((.........)))))))...)))((...(((((..((....))..)))))..)).... ( -40.00) >Bclau.0 218057 120 + 4303871/1320-1440 UUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAAGCCGGAAUUGCUAGUAAUCGCGGAUCAGCAUGCCGCGGUGAAUACGUUCCCGGGUCUUGUACACACCGCCCGUCACACCA ....((((((((..(((((((........))))))).....)).))))))...((((((((((.........)))))))...)))((...(((((..((....))..)))))..)).... ( -38.50) >Bsubt.0 90519 120 + 4214630/1320-1440 UUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGCUGGAAUCGCUAGUAAUCGCGGAUCAGCAUGCCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCA ..........((...((((((........))))))((((....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..))))))))).)). ( -42.70) >Blich.0 95327 120 + 4222334/1320-1440 UUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGCUGGAAUCGCUAGUAAUCGCGGAUCAGCAUGCCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCA ..........((...((((((........))))))((((....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..))))))))).)). ( -42.70) >Banth.0 82408 120 + 5227293/1320-1440 UUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCGCUAGUAAUCGCGGAUCAGCAUGCCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCA ..........((..(((((((........)))))))(((....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..))))))))..)). ( -39.30) >Bcere.0 82410 120 + 5224283/1320-1440 UUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCGCUAGUAAUCGCGGAUCAGCAUGCCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCA ..........((..(((((((........)))))))(((....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..))))))))..)). ( -39.30) >consensus UUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAAGCUGGAAUCGCUAGUAAUCGCGGAUCAGCAUGCCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCA ..........((..(((((((........)))))))(((....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..))))))))..)). (-41.07 = -39.38 + -1.69)
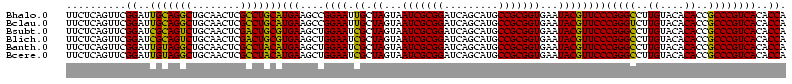
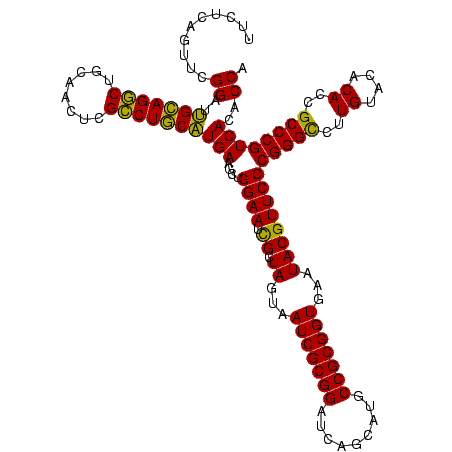
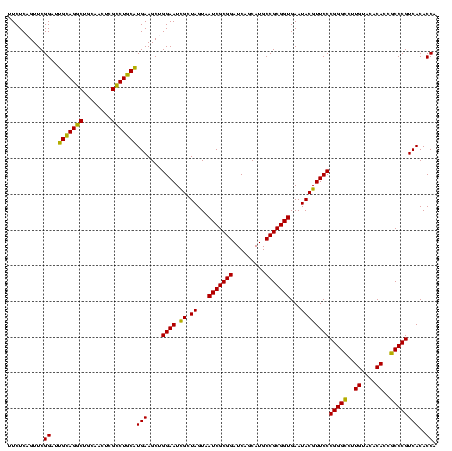
| Location | 656,180 – 656,299 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.71 |
| Mean single sequence MFE | -44.75 |
| Consensus MFE | -43.13 |
| Energy contribution | -43.05 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 119 + 4202352/1400-1520 AAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGGGGUAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGA ..........(((((..((....))..)))))...((((.(((..((((((..((((...((.((((..(((..((....-)).))).)))).))..))))..))))))..))).)))). ( -44.90) >Bclau.0 218057 119 + 4303871/1400-1520 AAUACGUUCCCGGGUCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGAGGCAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAAAUGAUUGGGGUGA ..........(((((..((....))..)))))...((((.(((..((((((..((((...((.((((..(((..((....-)).))).)))).))..))))..))))))..))).)))). ( -44.30) >Bsubt.0 90519 120 + 4214630/1400-1520 AAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUUAGGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGA ..........(((((..((....))..)))))...((((.(((..((((((..((((.....((((((.(((..((.....)).)))...)))))).))))..))))))..))).)))). ( -44.60) >Blich.0 95327 119 + 4222334/1400-1520 AAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUU-GGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGA ..........(((((..((....))..)))))...((((.(((..((((((..((((.....((((((.(((..((....-)).)))...)))))).))))..))))))..))).)))). ( -44.90) >Banth.0 82408 120 + 5227293/1400-1520 AAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGA ..........(((((..((....))..)))))...((((.(((..((((((..((((...((.((((..(((..((.....)).))).)))).))..))))..))))))..))).)))). ( -44.90) >Bcere.0 82410 119 + 5224283/1400-1520 AAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGGGGUAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGA ..........(((((..((....))..)))))...((((.(((..((((((..((((...((.((((..(((..((....-)).))).)))).))..))))..))))))..))).)))). ( -44.90) >consensus AAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUU_GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGA ..........(((((..((....))..)))))...((((.(((..((((((..((((...((.((((..(((..((.....)).))).)))).))..))))..))))))..))).)))). (-43.13 = -43.05 + -0.08)

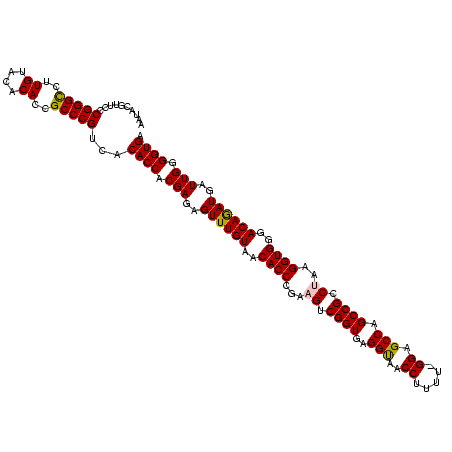
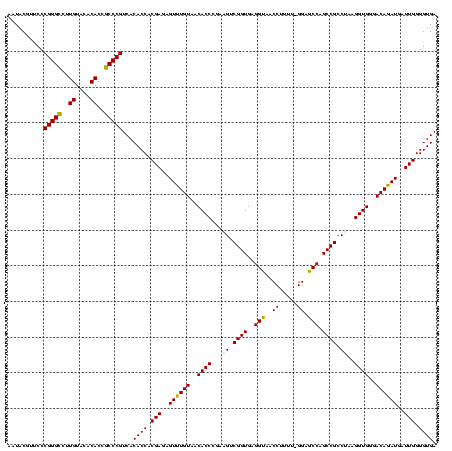
| Location | 656,180 – 656,299 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.99 |
| Mean single sequence MFE | -46.40 |
| Consensus MFE | -44.64 |
| Energy contribution | -44.70 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.91 |
| SVM RNA-class probability | 0.999961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 119 + 4202352/1440-1560 CGAGAGUUUGUAACACCCGAAGUCGGUGGGGUAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU (((..((((((..((((...((.((((..(((..((....-)).))).)))).))..))))..))))))..)))...................((((((((((....))))))))))... ( -46.30) >Bclau.0 218057 119 + 4303871/1440-1560 CGAGAGUUUGUAACACCCGAAGUCGGUGAGGCAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU (((..((((((..((((...((.((((..(((..((....-)).))).)))).))..))))..))))))..)))...................((((((((((....))))))))))... ( -47.20) >Bsubt.0 90519 120 + 4214630/1440-1560 CGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUUAGGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU (((..((((((..((((.....((((((.(((..((.....)).)))...)))))).))))..))))))..)))...................((((((((((....))))))))))... ( -46.00) >Blich.0 95327 119 + 4222334/1440-1560 CGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUU-GGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU (((..((((((..((((.....((((((.(((..((....-)).)))...)))))).))))..))))))..)))...................((((((((((....))))))))))... ( -46.30) >Banth.0 82408 120 + 5227293/1440-1560 CGAGAGUUUGUAACACCCGAAGUCGGUGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU (((..((((((..((((...((.((((..(((..((.....)).))).)))).))..))))..))))))..)))...................((((((((((....))))))))))... ( -46.30) >Bcere.0 82410 119 + 5224283/1440-1560 CGAGAGUUUGUAACACCCGAAGUCGGUGGGGUAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU (((..((((((..((((...((.((((..(((..((....-)).))).)))).))..))))..))))))..)))...................((((((((((....))))))))))... ( -46.30) >consensus CGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUU_GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU (((..((((((..((((...((.((((..(((..((.....)).))).)))).))..))))..))))))..)))...................((((((((((....))))))))))... (-44.64 = -44.70 + 0.06)

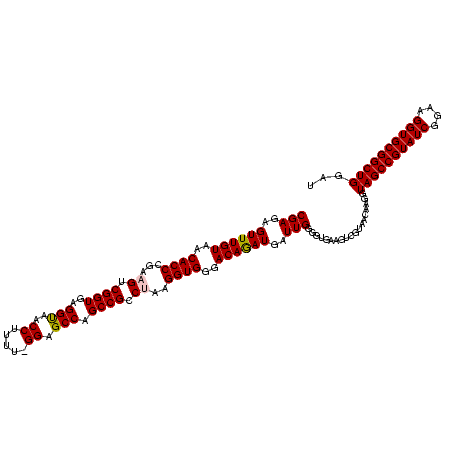
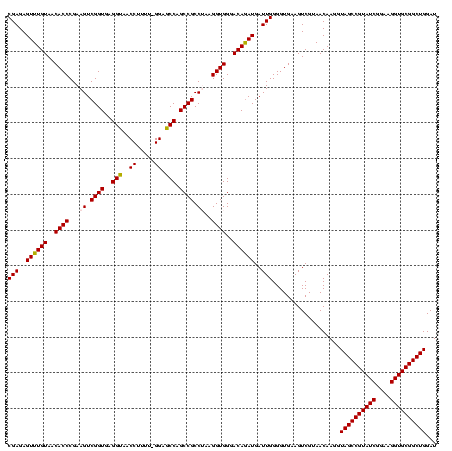
| Location | 656,180 – 656,299 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.34 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -37.82 |
| Energy contribution | -37.82 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 119 + 4202352/1480-1600 -GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUACGGAGUUUUUAACUCUAGUCGACCAUU -((......(((((...)))))(((....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))...(((((.....))))).)))..))... ( -45.60) >Bclau.0 218057 118 + 4303871/1480-1600 -GGAGCCAGCCGCCUAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGUUUUUA-CUCUAGUCGAUGUAU -........(((((...)))))(((....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..((((((....)-))))))))....... ( -45.20) >Bsubt.0 90519 118 + 4214630/1480-1600 AGGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUUUA--CGGAAUAUAAGACC .((..((..(((((...))))).......((..(((((((.((....))....((((((((((....))))))))))..))))).))..))...))((((((..--..))))))....)) ( -39.70) >Blich.0 95327 117 + 4222334/1480-1600 -GGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUA--CGGAAUAUAAGACC -((..((..(((((...))))).......((..(((((((.((....))....((((((((((....))))))))))..))))).))..))...))(((((...--...)))))....)) ( -38.40) >Banth.0 82408 116 + 5227293/1480-1600 UGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA----UGAACGCUGUUC ...(((...(((((...)))))..((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..)).........----.....))).... ( -40.10) >Bcere.0 82410 115 + 5224283/1480-1600 -GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA----UGAACGCUGUUC -..(((...(((((...)))))..((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..)).........----.....))).... ( -40.10) >consensus _GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAUUUUA____UAAUCGAUGAUC .....((..(((((...))))).......((..(((((((.((....))....((((((((((....))))))))))..))))).))..))...))........................ (-37.82 = -37.82 + 0.00)
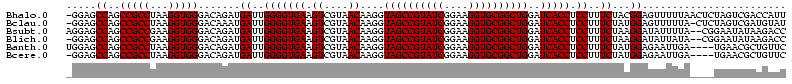
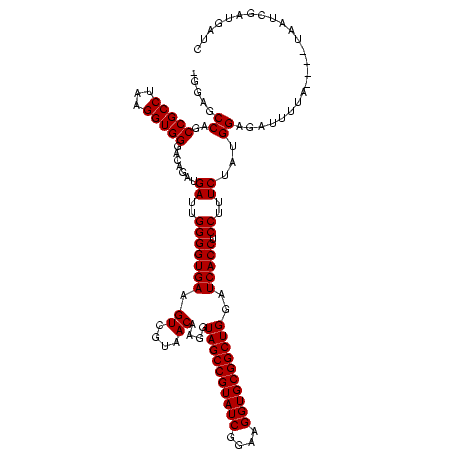

| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.07 |
| Mean single sequence MFE | -45.32 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.93 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/1506-1626 GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUACGGAGUUUUUAACUCUAGUCGACCAUUCGCAGAUUCAUCUGCGGAAGGACGCU ..((((((((((((..(((((..((((.(((((((((....)))))))))((....)).))))..)))))..))....)))))))))).((.((((((((....)))))))).))..... ( -50.20) >Bclau.0 218057 115 + 4303871/1506-1626 AAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGUUUUUA-CUCUAGUCGAUGUAU----GACUUUCUGGUCAUAUAGCGCU ..((((((((((((((...........((((((((((....))))))))))......((((.......)))).)))))-))))))))).(((((----((((....)))))))))..... ( -47.70) >Bsubt.0 90519 117 + 4214630/1506-1626 GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUUUA--CGGAAUAUAAGACCUUG-GGUCUUAUAAACAGAACGUUCC .......((..((...((((((...((((((((((((....))))))))).....)))((((.....))))....)))--)))..(((((((((...-))))))))).......))..)) ( -40.50) >Blich.0 95327 118 + 4222334/1506-1626 GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUA--CGGAAUAUAAGACCUUGCGGUCUUAUAGACAGGUGCGUUU ...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))...........((--(.(..(((((((((....)))))))))..)..)))..... ( -41.90) >Banth.0 82408 116 + 5227293/1506-1626 GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA----UGAACGCUGUUCAUCAAUAUAAGUUUCCGUGUUUCGUU ...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))(((((((((((((----(((((...)))))))))).....))))))))....... ( -45.80) >Bcere.0 82410 116 + 5224283/1506-1626 GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA----UGAACGCUGUUCAUCAAUAUAAGUUUCCGUGUUUCGUU ...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))(((((((((((((----(((((...)))))))))).....))))))))....... ( -45.80) >consensus GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAUUUUA____UAAUCGAUGAUCAUCAGAUUUAAUUGCCAUAUGGCGCU ...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))....................................................... (-29.70 = -29.70 + -0.00)

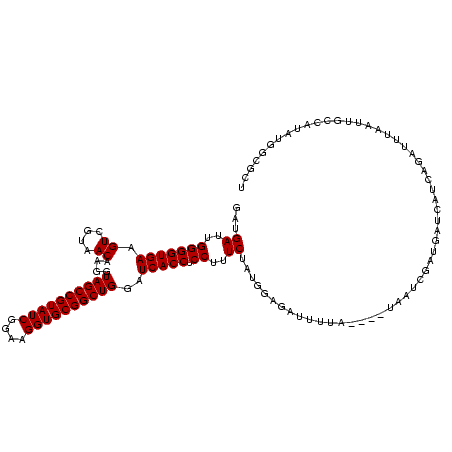
| Location | 656,180 – 656,300 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.07 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -15.20 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 656180 120 + 4202352/1506-1626 AGCGUCCUUCCGCAGAUGAAUCUGCGAAUGGUCGACUAGAGUUAAAAACUCCGUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC ((((.((...((((((....))))))...)).)).)).((((.....))))(((((.((((.((....))(((((............))))).)))).)))))................. ( -33.20) >Bclau.0 218057 115 + 4303871/1506-1626 AGCGCUAUAUGACCAGAAAGUC----AUACAUCGACUAGAG-UAAAAACUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUU ((((...((((((......)))----)))...)).)).(((-(....))))....((..((.(((((...(((((............))))).....((....)).)))))))..))... ( -24.10) >Bsubt.0 90519 117 + 4214630/1506-1626 GGAACGUUCUGUUUAUAAGACC-CAAGGUCUUAUAUUCCG--UAAAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC (((...(((.(..(((((((((-...)))))))))..).)--.))....)))...((..((.(((((...(((((............))))).....((....)).)))))))..))... ( -27.10) >Blich.0 95327 118 + 4222334/1506-1626 AAACGCACCUGUCUAUAAGACCGCAAGGUCUUAUAUUCCG--UAUAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC ..........((((((((((((....)))))))))..(((--(((....((((.....))))((((.........)))).....))))))............)))............... ( -30.90) >Banth.0 82408 116 + 5227293/1506-1626 AACGAAACACGGAAACUUAUAUUGAUGAACAGCGUUCA----UCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC ..((.((((.(((.......((((((((((...)))))----)))))(((((.......)))))...)))(((((............)))))....)))).))................. ( -28.00) >Bcere.0 82410 116 + 5224283/1506-1626 AACGAAACACGGAAACUUAUAUUGAUGAACAGCGUUCA----UCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC ..((.((((.(((.......((((((((((...)))))----)))))(((((.......)))))...)))(((((............)))))....)))).))................. ( -28.00) >consensus AACGAAACACGGCAACAAAAACUGAAGAACAUCGAUUA____UAAAAUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC .......................................................((..((.(((((...(((((............))))).....((....)).)))))))..))... (-15.20 = -15.20 + 0.00)
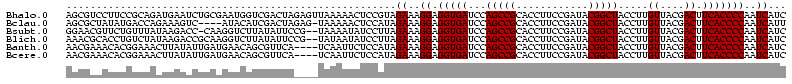
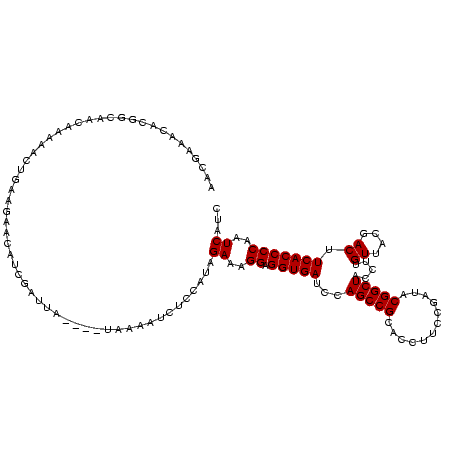
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:16:39 2006