| Sequence ID | Bcere.0 |
|---|---|
| Location | 5,211,703 – 5,211,791 |
| Length | 88 |
| Max. P | 0.573289 |

| Location | 5,211,703 – 5,211,791 |
|---|---|
| Length | 88 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 83.99 |
| Mean single sequence MFE | -19.48 |
| Consensus MFE | -11.25 |
| Energy contribution | -11.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
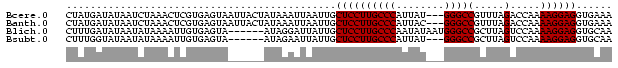
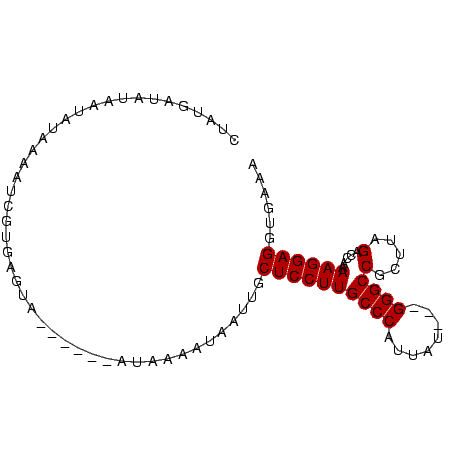
>Bcere.0 5211703 88 + 5224283 UUUCACCUCCUUUUGGUCUAAACGGCCC---AUAAUGGGCAAGGAGCAAUUAAUUUAUAGUAAUUACUCACGAGUUUAGAUUAUAUCAUAG .............((((((((((.((((---.....))))...(((.(((((........))))).)))....))))))))))........ ( -22.20) >Banth.0 5214712 88 + 5227293 UUUCACCUCCUUUUGGUCUAAACGGCCC---GUAAUGGGCAAGGAGCAAUUAAUUUAUAGUAAUUACUCACGAGUUUAGAUUAUAUCAUAG .............((((((((((.((((---.....))))...(((.(((((........))))).)))....))))))))))........ ( -21.50) >Blich.0 4207817 85 + 4222334 UUGCACCUCCUUUUGGACUAAGCGGCCCAUUAUAUUGGGCAAGGAGCAAUAAUCCUAU------UACUCACAAUUUUAUAUUAUAUCAAAG ((((.(((((....))).......(((((......)))))..)).)))).........------........................... ( -17.90) >Bsubt.0 4198760 82 + 4214630 UUGCACCUCCUUUUGGACUAAGCGGCCC---AUAAUGGGCAAGGAGCAAUAAUUCUAU------UACUCACAAUUUUAUAUUAUACCAAAG ((((.(((((....))).......((((---.....))))..)).)))).........------........................... ( -16.30) >consensus UUGCACCUCCUUUUGGACUAAACGGCCC___AUAAUGGGCAAGGAGCAAUAAAUCUAU______UACUCACAAGUUUAGAUUAUAUCAAAG ......((((((...(......).((((........))))))))))............................................. (-11.25 = -11.25 + 0.00)

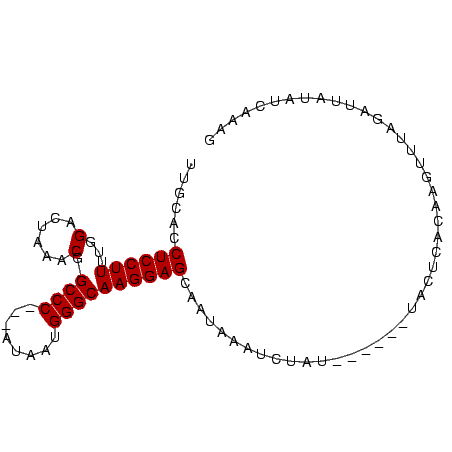
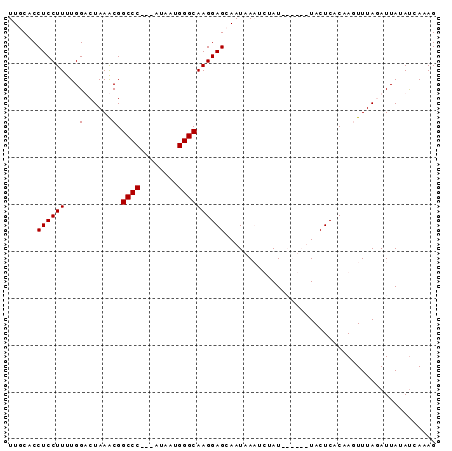
| Location | 5,211,703 – 5,211,791 |
|---|---|
| Length | 88 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 83.99 |
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -12.27 |
| Energy contribution | -12.28 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5211703 88 + 5224283 CUAUGAUAUAAUCUAAACUCGUGAGUAAUUACUAUAAAUUAAUUGCUCCUUGCCCAUUAU---GGGCCGUUUAGACCAAAAGGAGGUGAAA ...........(((((((..(.(((((((((........))))))))))..((((.....---)))).)))))))((....))........ ( -22.60) >Banth.0 5214712 88 + 5227293 CUAUGAUAUAAUCUAAACUCGUGAGUAAUUACUAUAAAUUAAUUGCUCCUUGCCCAUUAC---GGGCCGUUUAGACCAAAAGGAGGUGAAA ...........(((((((..(.(((((((((........))))))))))..((((.....---)))).)))))))((....))........ ( -22.30) >Blich.0 4207817 85 + 4222334 CUUUGAUAUAAUAUAAAAUUGUGAGUA------AUAGGAUUAUUGCUCCUUGCCCAAUAUAAUGGGCCGCUUAGUCCAAAAGGAGGUGCAA ......................(((((------(((....))))))))...(((((......))))).((....(((....)))...)).. ( -18.40) >Bsubt.0 4198760 82 + 4214630 CUUUGGUAUAAUAUAAAAUUGUGAGUA------AUAGAAUUAUUGCUCCUUGCCCAUUAU---GGGCCGCUUAGUCCAAAAGGAGGUGCAA ......................(((((------(((....))))))))...((((.....---)))).((....(((....)))...)).. ( -17.60) >consensus CUAUGAUAUAAUAUAAAAUCGUGAGUA______AUAAAAUAAUUGCUCCUUGCCCAUUAU___GGGCCGCUUAGACCAAAAGGAGGUGAAA .............................................((((((((((........))))(.....).....))))))...... (-12.27 = -12.28 + 0.00)
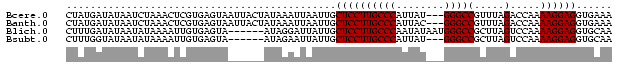
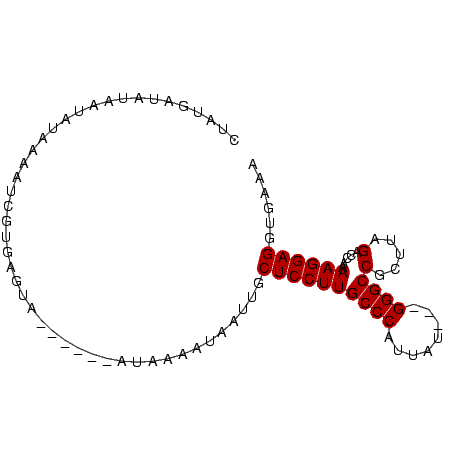
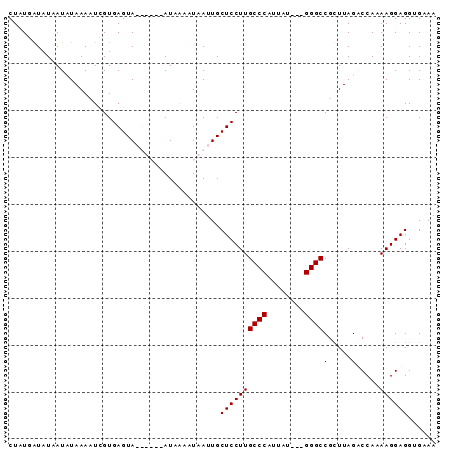
| Location | 5,211,703 – 5,211,791 |
|---|---|
| Length | 88 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 83.99 |
| Mean single sequence MFE | -19.48 |
| Consensus MFE | -11.25 |
| Energy contribution | -11.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5211703 88 + 5224283/0-91 UUUCACCUCCUUUUGGUCUAAACGGCCC---AUAAUGGGCAAGGAGCAAUUAAUUUAUAGUAAUUACUCACGAGUUUAGAUUAUAUCAUAG .............((((((((((.((((---.....))))...(((.(((((........))))).)))....))))))))))........ ( -22.20) >Banth.0 5214712 88 + 5227293/0-91 UUUCACCUCCUUUUGGUCUAAACGGCCC---GUAAUGGGCAAGGAGCAAUUAAUUUAUAGUAAUUACUCACGAGUUUAGAUUAUAUCAUAG .............((((((((((.((((---.....))))...(((.(((((........))))).)))....))))))))))........ ( -21.50) >Blich.0 4207817 85 + 4222334/0-91 UUGCACCUCCUUUUGGACUAAGCGGCCCAUUAUAUUGGGCAAGGAGCAAUAAUCCUAU------UACUCACAAUUUUAUAUUAUAUCAAAG ((((.(((((....))).......(((((......)))))..)).)))).........------........................... ( -17.90) >Bsubt.0 4198760 82 + 4214630/0-91 UUGCACCUCCUUUUGGACUAAGCGGCCC---AUAAUGGGCAAGGAGCAAUAAUUCUAU------UACUCACAAUUUUAUAUUAUACCAAAG ((((.(((((....))).......((((---.....))))..)).)))).........------........................... ( -16.30) >consensus UUGCACCUCCUUUUGGACUAAACGGCCC___AUAAUGGGCAAGGAGCAAUAAAUCUAU______UACUCACAAGUUUAGAUUAUAUCAAAG ......((((((...(......).((((........))))))))))............................................. (-11.25 = -11.25 + 0.00)

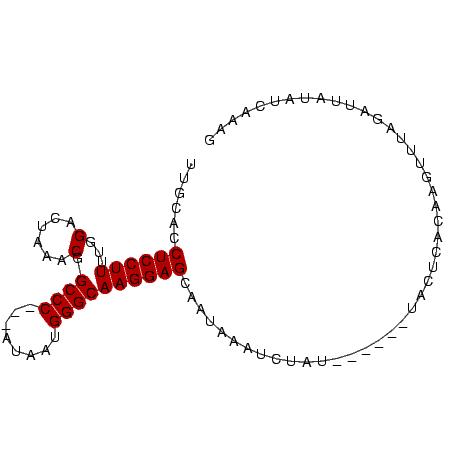
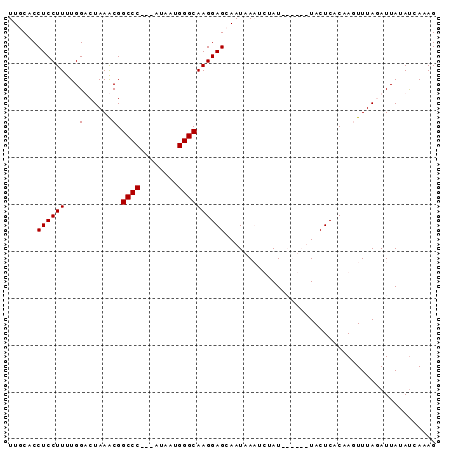
| Location | 5,211,703 – 5,211,791 |
|---|---|
| Length | 88 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 83.99 |
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -12.27 |
| Energy contribution | -12.28 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5211703 88 + 5224283/0-91 CUAUGAUAUAAUCUAAACUCGUGAGUAAUUACUAUAAAUUAAUUGCUCCUUGCCCAUUAU---GGGCCGUUUAGACCAAAAGGAGGUGAAA ...........(((((((..(.(((((((((........))))))))))..((((.....---)))).)))))))((....))........ ( -22.60) >Banth.0 5214712 88 + 5227293/0-91 CUAUGAUAUAAUCUAAACUCGUGAGUAAUUACUAUAAAUUAAUUGCUCCUUGCCCAUUAC---GGGCCGUUUAGACCAAAAGGAGGUGAAA ...........(((((((..(.(((((((((........))))))))))..((((.....---)))).)))))))((....))........ ( -22.30) >Blich.0 4207817 85 + 4222334/0-91 CUUUGAUAUAAUAUAAAAUUGUGAGUA------AUAGGAUUAUUGCUCCUUGCCCAAUAUAAUGGGCCGCUUAGUCCAAAAGGAGGUGCAA ......................(((((------(((....))))))))...(((((......))))).((....(((....)))...)).. ( -18.40) >Bsubt.0 4198760 82 + 4214630/0-91 CUUUGGUAUAAUAUAAAAUUGUGAGUA------AUAGAAUUAUUGCUCCUUGCCCAUUAU---GGGCCGCUUAGUCCAAAAGGAGGUGCAA ......................(((((------(((....))))))))...((((.....---)))).((....(((....)))...)).. ( -17.60) >consensus CUAUGAUAUAAUAUAAAAUCGUGAGUA______AUAAAAUAAUUGCUCCUUGCCCAUUAU___GGGCCGCUUAGACCAAAAGGAGGUGAAA .............................................((((((((((........))))(.....).....))))))...... (-12.27 = -12.28 + 0.00)
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:26:20 2006