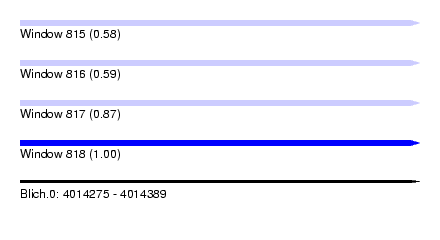
| Sequence ID | Blich.0 |
|---|---|
| Location | 4,014,275 – 4,014,389 |
| Length | 114 |
| Max. P | 0.995082 |

| Location | 4,014,275 – 4,014,389 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 60.17 |
| Mean single sequence MFE | -43.13 |
| Consensus MFE | -8.43 |
| Energy contribution | -11.43 |
| Covariance contribution | 3.01 |
| Combinations/Pair | 1.18 |
| Mean z-score | -4.45 |
| Structure conservation index | 0.20 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 4014275 114 + 4222334/0-120 UGAAACCCGGCAACCGCUGUCUAUGACAGAAUG--GUGCUAAAUCCUUAAGAGCAUGUUCGUGCUCU-UGAAGAUAAGGAGGAGAUUGAAUAUAUCAAGCUCUUCCUUAUC---GGGAAG .....(((((((.((.(((((...)))))...)--)))))......((((((((((....)))))))-))).((((((((((((.((((.....)))).))))))))))))---)))... ( -50.80) >Bsubt.0 3998216 112 + 4214630/0-120 CGAUACCCGGCAACCGCUGUUUA--ACAGAAUG--GUGCUAAAUCCUUUAGAGCA-AUGAUUGCUCU-UGAAGAUAAGGUUGAGAUUG--UCACGCAAGCUCUUCCUUAUCCAAAGGAAG ........((((.((.(((....--.)))...)--)))))...((((((((((((-.....))))))-....(((((((..(((.(((--.....))).)))..))))))).)))))).. ( -35.90) >Banth.0 1375383 115 + 5227293/0-120 CGAUGCCUCGGCAGCGGACUCGAUUUUAGAGUGCUGUGCCAAAUCCAGCA-AGCAUGUGCUUGAAAGAUGAGAAGAGCGUUUCUUAUAGAUGUAUAAGACCUCUUCUCGUU----GGAAG .........((((.(((((((.......)))).)))))))...(((..((-(((....)))))...(((((((((((.((((..(((....)))..)))))))))))))))----))).. ( -42.70) >consensus CGAUACCCGGCAACCGCUGUCUAU_ACAGAAUG__GUGCUAAAUCCUUAAGAGCAUGUGCUUGCUCU_UGAAGAUAAGGUUGAGAUUG_AUAUAUCAAGCUCUUCCUUAUC____GGAAG ...........................................(((....(((((......)))))......(((((((..(((.((((.....)))).)))..)))))))....))).. ( -8.43 = -11.43 + 3.01)

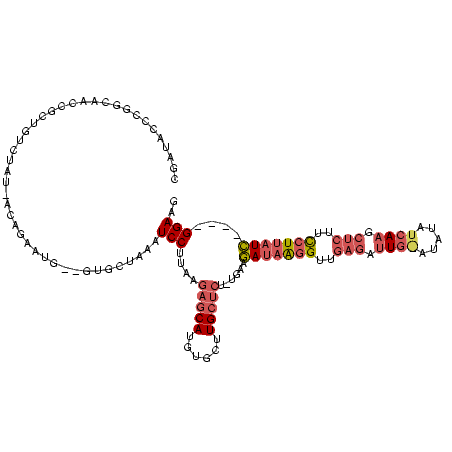
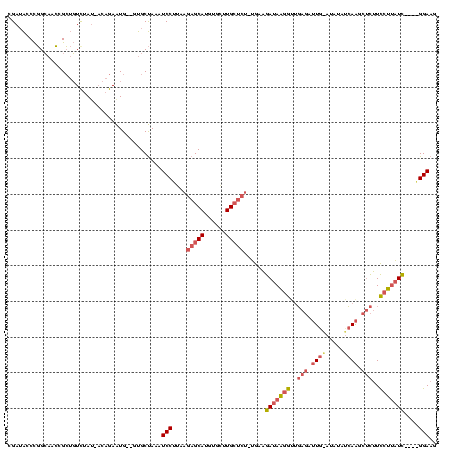
| Location | 4,014,275 – 4,014,389 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 60.17 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -8.70 |
| Energy contribution | -8.50 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.45 |
| Mean z-score | -3.79 |
| Structure conservation index | 0.23 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 4014275 114 + 4222334/0-120 CUUCCC---GAUAAGGAAGAGCUUGAUAUAUUCAAUCUCCUCCUUAUCUUCA-AGAGCACGAACAUGCUCUUAAGGAUUUAGCAC--CAUUCUGUCAUAGACAGCGGUUGCCGGGUUUCA ...(((---(.((((((.(((.((((.....)))).))).))))))((((.(-((((((......))))))).))))....((((--(...(((((...))))).)).)))))))..... ( -39.80) >Bsubt.0 3998216 112 + 4214630/0-120 CUUCCUUUGGAUAAGGAAGAGCUUGCGUGA--CAAUCUCAACCUUAUCUUCA-AGAGCAAUCAU-UGCUCUAAAGGAUUUAGCAC--CAUUCUGU--UAAACAGCGGUUGCCGGGUAUCG ...(((((((((((((..(((.(((.....--))).)))..))))))))...-((((((.....-)))))))))))(((..((((--(...(((.--....))).)).)))..))).... ( -34.30) >Banth.0 1375383 115 + 5227293/0-120 CUUCC----AACGAGAAGAGGUCUUAUACAUCUAUAAGAAACGCUCUUCUCAUCUUUCAAGCACAUGCU-UGCUGGAUUUGGCACAGCACUCUAAAAUCGAGUCCGCUGCCGAGGCAUCG ..(((----(..((((((((.(((((((....)))))))....))))))))......(((((....)))-)).))))(((((((..(.((((.......)))).)..)))))))...... ( -38.40) >consensus CUUCC____GAUAAGGAAGAGCUUGAUACAU_CAAUCUCAACCUUAUCUUCA_AGAGCAAGCACAUGCUCUAAAGGAUUUAGCAC__CAUUCUGU_AUAGACAGCGGUUGCCGGGUAUCG (((((.........))))).(((((.....................(((....((((((......))))))...)))......................(((....)))..))))).... ( -8.70 = -8.50 + -0.20)

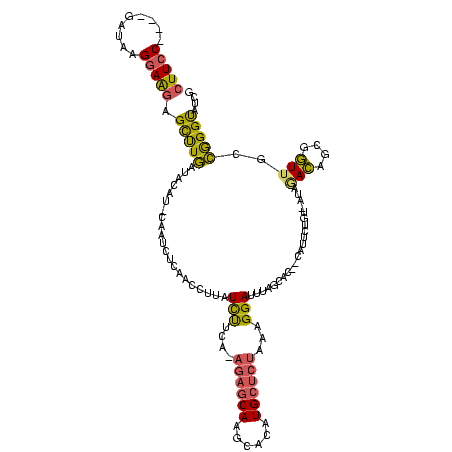
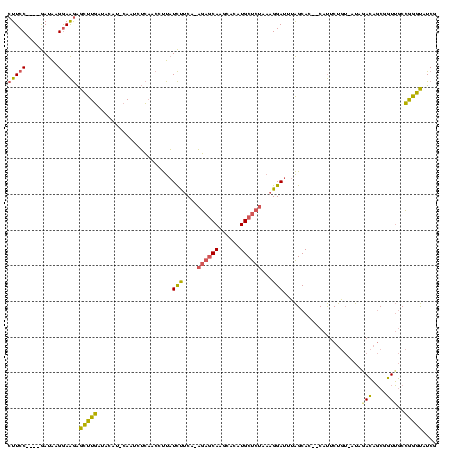
| Location | 4,014,275 – 4,014,389 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.02 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -10.77 |
| Energy contribution | -12.67 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.90 |
| Structure conservation index | 0.28 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 4014275 114 + 4222334/14-134 CGCUGUCUAUGACAGAAUG--GUGCUAAAUCCUUAAGAGCAUGUUCGUGCUCU-UGAAGAUAAGGAGGAGAUUGAAUAUAUCAAGCUCUUCCUUAUC---GGGAAGAGCUUUUUUAUGUC ..(((((...)))))....--..(((...(((((((((((((....)))))))-))).((((((((((((.((((.....)))).))))))))))))---.)))..)))........... ( -45.10) >Bsubt.0 3998216 112 + 4214630/14-134 CGCUGUUUA--ACAGAAUG--GUGCUAAAUCCUUUAGAGCA-AUGAUUGCUCU-UGAAGAUAAGGUUGAGAUUG--UCACGCAAGCUCUUCCUUAUCCAAAGGAAGAGCUUUUUUAUAUU .(((((((.--...)))))--))(((...((((((((((((-.....))))))-....(((((((..(((.(((--.....))).)))..))))))).))))))..)))........... ( -34.00) >Banth.0 1375383 115 + 5227293/14-134 CGGACUCGAUUUUAGAGUGCUGUGCCAAAUCCAGCA-AGCAUGUGCUUGAAAGAUGAGAAGAGCGUUUCUUAUAGAUGUAUAAGACCUCUUCUCGUU----GGAAGAGGUCUUUUGUUAU (((((((.......)))).))).(((...(((..((-(((....)))))...(((((((((((.((((..(((....)))..)))))))))))))))----)))...))).......... ( -36.40) >consensus CGCUGUCUAU_ACAGAAUG__GUGCUAAAUCCUUAAGAGCAUGUGCUUGCUCU_UGAAGAUAAGGUUGAGAUUG_AUAUAUCAAGCUCUUCCUUAUC____GGAAGAGCUUUUUUAUAUU ....................................(((((......)))))..............................(((((((((((.......)))))))))))......... (-10.77 = -12.67 + 1.90)

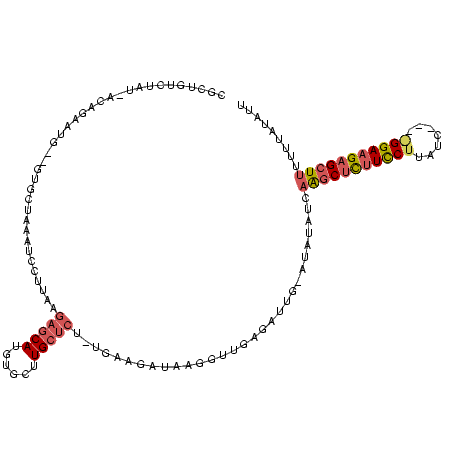
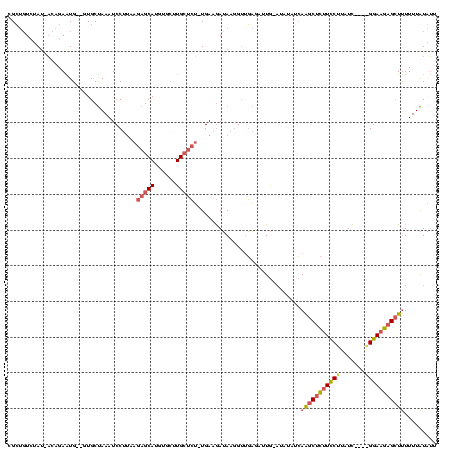
| Location | 4,014,275 – 4,014,389 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 61.02 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -12.54 |
| Energy contribution | -13.67 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.26 |
| Mean z-score | -4.76 |
| Structure conservation index | 0.36 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 4014275 114 + 4222334/14-134 GACAUAAAAAAGCUCUUCCC---GAUAAGGAAGAGCUUGAUAUAUUCAAUCUCCUCCUUAUCUUCA-AGAGCACGAACAUGCUCUUAAGGAUUUAGCAC--CAUUCUGUCAUAGACAGCG ...........(((..(((.---((((((((.(((.((((.....)))).))).))))))))...(-((((((......)))))))..)))...)))..--....(((((...))))).. ( -36.50) >Bsubt.0 3998216 112 + 4214630/14-134 AAUAUAAAAAAGCUCUUCCUUUGGAUAAGGAAGAGCUUGCGUGA--CAAUCUCAACCUUAUCUUCA-AGAGCAAUCAU-UGCUCUAAAGGAUUUAGCAC--CAUUCUGU--UAAACAGCG .........((((((((((((.....))))))))))))((.(((--((........((.(((((..-((((((.....-))))))..)))))..))...--.....)))--))....)). ( -34.59) >Banth.0 1375383 115 + 5227293/14-134 AUAACAAAAGACCUCUUCC----AACGAGAAGAGGUCUUAUACAUCUAUAAGAAACGCUCUUCUCAUCUUUCAAGCACAUGCU-UGCUGGAUUUGGCACAGCACUCUAAAAUCGAGUCCG .........(.((...(((----(..((((((((.(((((((....)))))))....))))))))......(((((....)))-)).))))...)))...(.((((.......)))).). ( -32.50) >consensus AAAAUAAAAAAGCUCUUCC____GAUAAGGAAGAGCUUGAUACAU_CAAUCUCAACCUUAUCUUCA_AGAGCAAGCACAUGCUCUAAAGGAUUUAGCAC__CAUUCUGU_AUAGACAGCG ....((((.((((((((((.........))))))))))......................(((....((((((......))))))...)))))))......................... (-12.54 = -13.67 + 1.12)

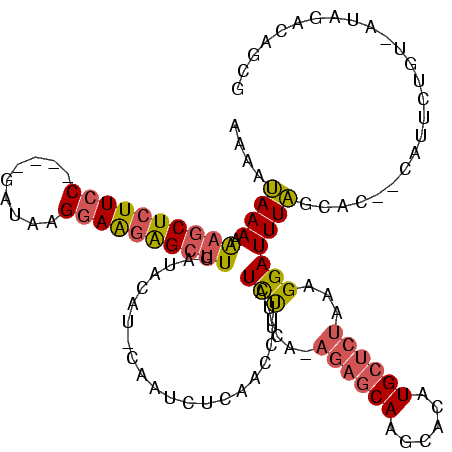
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:26:17 2006