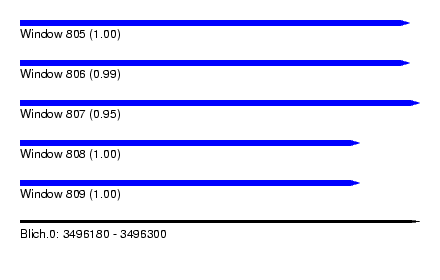
| Sequence ID | Blich.0 |
|---|---|
| Location | 3,496,180 – 3,496,300 |
| Length | 120 |
| Max. P | 0.999973 |

| Location | 3,496,180 – 3,496,297 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.67 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -18.73 |
| Energy contribution | -20.60 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.47 |
| Structure conservation index | 0.59 |
| SVM decision value | 5.08 |
| SVM RNA-class probability | 0.999973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 3496180 117 + 4222334/0-120 AAAAUAAAAAA---GCUCACCGUCUCCCUCUUCAAUUUGAAGAUAAAGAGACGAUGAGCUUUGAAGCUUACCGCGGUACCACUCUUUUUGAUCCUAUACCAAGAAUCCAGCUUUGCGAAG ........(((---(((((.((((((..(((((.....)))))....)))))).))))))))(((((((((....)))......((((((.........))))))...))))))...... ( -32.70) >Bsubt.0 3490312 111 + 4214630/0-120 AAAACAAAAAA---GCCCGCCGUCUCCUACCUAAUCUAAUAGAUAAAGAGACGACGAGCCUUCG-GCUGACCGCGGUACCACUCUUGUUGAUUCUUCAUGAAGAAUCCUGCUUUU----- .......((((---((((((((((((.((.(((......))).))..))))))...((((...)-)))....)))).............(((((((....)))))))..))))))----- ( -28.90) >Bcere.0 3070925 104 - 5224283/0-120 AAAAUAAAAAAAUGGCUUAGCGUCUCU-------------AUAUAAAGAGACGCUAAGCCAUUU-GCUUAACGCGGUACCACUCUUAUUGAUUC--UAUUAUGAAUCCACCUUAGUAGUA .........((((((((((((((((((-------------......))))))))))))))))))-((.....))..(((.(((......(((((--......)))))......))).))) ( -38.00) >Banth.0 3158000 105 - 5227293/0-120 AAAAUAAAAAA-UGGCUCAGCAUCUCUUU-----------AUAUAAAGAGACGCUAAGCCAUUU-GCUUAACGCGGUACCACCCUUAUUGAUUC--CAUUAUGAAUCCACCUUAGUAGUA ..(((((.(((-(((((.(((.(((((((-----------....))))))).))).))))))))-((.....))((.....)).)))))(((((--......)))))............. ( -27.40) >consensus AAAAUAAAAAA___GCUCACCGUCUCCUU___________AGAUAAAGAGACGAUAAGCCAUUU_GCUUAACGCGGUACCACUCUUAUUGAUUC__CAUUAAGAAUCCACCUUAGUAGUA ..(((((......(((((((((((((.....................))))))))))))).....((.....))..........)))))((((((......))))))............. (-18.73 = -20.60 + 1.87)

| Location | 3,496,180 – 3,496,297 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.67 |
| Mean single sequence MFE | -45.20 |
| Consensus MFE | -32.84 |
| Energy contribution | -31.16 |
| Covariance contribution | -1.69 |
| Combinations/Pair | 1.37 |
| Mean z-score | -6.51 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 3496180 117 + 4222334/0-120 CUUCGCAAAGCUGGAUUCUUGGUAUAGGAUCAAAAAGAGUGGUACCGCGGUAAGCUUCAAAGCUCAUCGUCUCUUUAUCUUCAAAUUGAAGAGGGAGACGGUGAGC---UUUUUUAUUUU .......(((((.((((((......)))))).......(((....)))....))))).((((((((((((((((...(((((.....))))).)))))))))))))---)))........ ( -44.20) >Bsubt.0 3490312 111 + 4214630/0-120 -----AAAAGCAGGAUUCUUCAUGAAGAAUCAACAAGAGUGGUACCGCGGUCAGC-CGAAGGCUCGUCGUCUCUUUAUCUAUUAGAUUAGGUAGGAGACGGCGGGC---UUUUUUGUUUU -----.((((((((((((((....))))))).......(((....)))((....)-)((((((((((((((((((.(((((......)))))))))))))))))))---))))))))))) ( -46.50) >Bcere.0 3070925 104 - 5224283/0-120 UACUACUAAGGUGGAUUCAUAAUA--GAAUCAAUAAGAGUGGUACCGCGUUAAGC-AAAUGGCUUAGCGUCUCUUUAUAU-------------AGAGACGCUAAGCCAUUUUUUUAUUUU (((((((...((.(((((......--))))).))...)))))))..((.....))-((((((((((((((((((......-------------))))))))))))))))))......... ( -45.30) >Banth.0 3158000 105 - 5227293/0-120 UACUACUAAGGUGGAUUCAUAAUG--GAAUCAAUAAGGGUGGUACCGCGUUAAGC-AAAUGGCUUAGCGUCUCUUUAUAU-----------AAAGAGAUGCUGAGCCA-UUUUUUAUUUU (((((((...((.(((((......--))))).))...)))))))..((.....))-((((((((((((((((((((....-----------)))))))))))))))))-)))........ ( -44.80) >consensus UACUACAAAGCUGGAUUCAUAAUA__GAAUCAAUAAGAGUGGUACCGCGGUAAGC_AAAAGGCUCAGCGUCUCUUUAUAU___________AAAGAGACGCUGAGC___UUUUUUAUUUU .............(((((........)))))((((((((((....)))............(((((((((((((((.................)))))))))))))))....))))))).. (-32.84 = -31.16 + -1.69)

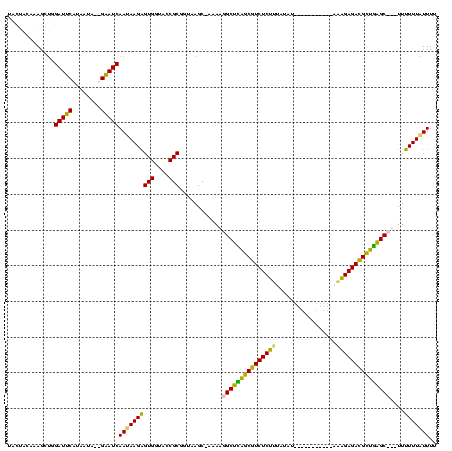
| Location | 3,496,180 – 3,496,300 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.97 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -18.21 |
| Energy contribution | -17.90 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 3496180 120 + 4222334/40-160 CGAUUAACCUUCGUUAACAGGUUUGCACAGCAUGGCUGUGCUUCGCAAAGCUGGAUUCUUGGUAUAGGAUCAAAAAGAGUGGUACCGCGGUAAGCUUCAAAGCUCAUCGUCUCUUUAUCU ...(((((....)))))(((.(((((..(((((....)))))..))))).)))((((((......))))))..((((((.(....)(((((.((((....)))).))))))))))).... ( -34.70) >Bsubt.0 3490312 96 + 4214630/40-160 CGGCUAACCUUCGUU-ACAGGUC----------------------AAAAGCAGGAUUCUUCAUGAAGAAUCAACAAGAGUGGUACCGCGGUCAGC-CGAAGGCUCGUCGUCUCUUUAUCU (((((.(((......-.......----------------------........(((((((....))))))).......(((....)))))).)))-)).((((.....))))........ ( -25.00) >Bcere.0 3070925 114 - 5224283/40-160 CGGUUAACCUUCGAUAUUAGGUUUGUA---AUCAAACGAUUACUACUAAGGUGGAUUCAUAAUA--GAAUCAAUAAGAGUGGUACCGCGUUAAGC-AAAUGGCUUAGCGUCUCUUUAUAU .(((..((((((....((((....(((---(((....))))))..))))....(((((......--))))).....))).))))))(((((((((-.....))))))))).......... ( -31.30) >Banth.0 3158000 114 - 5227293/40-160 CGGUUAACCUUCGAUAUUAGGUUUGUA---AUCGAACGAUUACUACUAAGGUGGAUUCAUAAUG--GAAUCAAUAAGGGUGGUACCGCGUUAAGC-AAAUGGCUUAGCGUCUCUUUAUAU .(((..(((((((...((((....(((---(((....))))))..))))..))(((((......--)))))....)))))...)))(((((((((-.....))))))))).......... ( -33.70) >consensus CGGUUAACCUUCGAUAACAGGUUUGUA___AUCGAACGAUUACUACAAAGCUGGAUUCAUAAUA__GAAUCAAUAAGAGUGGUACCGCGGUAAGC_AAAAGGCUCAGCGUCUCUUUAUAU .....(((((........)))))..............................(((((........)))))...(((((.(....)(((((((((......))))))))))))))..... (-18.21 = -17.90 + -0.31)

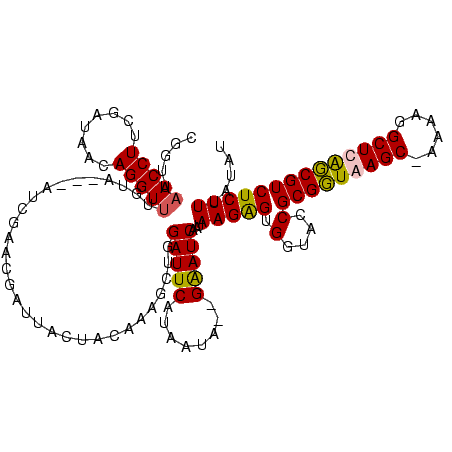
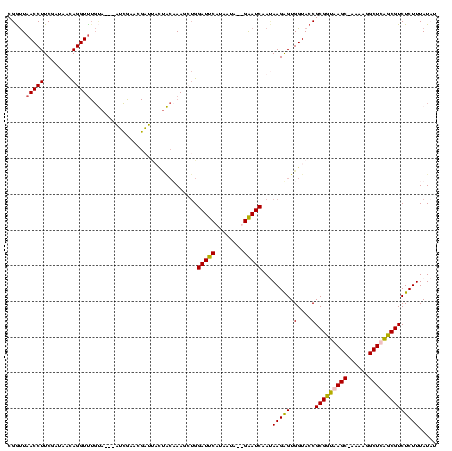
| Location | 3,496,180 – 3,496,282 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.05 |
| Mean single sequence MFE | -20.17 |
| Consensus MFE | -21.14 |
| Energy contribution | -16.07 |
| Covariance contribution | -5.06 |
| Combinations/Pair | 1.66 |
| Mean z-score | -0.71 |
| Structure conservation index | 1.05 |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 3496180 102 + 4222334/200-320 UCCACUCCCGGGCGAGUUCAAAGGUUCAUUGGUCUGCGUCUCACCUACCCGCAGCUCUCUUUGCCAAUGCCGAUCUUUUACUGCUCCCGUUCAUUGUGUUUU------------------ ..(((...((((.((((..(((((((((((((((((((...........)))))........)))))))..))))))).))).).))))......)))....------------------ ( -26.70) >Bsubt.0 3490312 120 + 4214630/200-320 ACCACUCCCGGGCGAGUUCAAAGAAGAAUCAGGCUGCGUCGCACCACCCCGCAGCUCUCUGUUCUGAUCACUUUCUUUUACUGCUCCCAAUCUAUGUGUUUUAGUUUUCUGAAAAACUCA .........(((.((((..(((((((.(((((((((((..(.....)..))))))........)))))...))))))).))).).))).......((.((((((....)))))).))... ( -27.40) >Bcere.0 3070925 114 - 5224283/200-320 ACUGCUCCUAGGCGAGUUCAGGAUGUCAUUUGACUAUGUCACACCAACCCAUAGCUCUCUGUACAAAU-AUACGACCUUACUACUCCUAUUCAACACAUUUUAAUAUUUAGAAUA----- ...(((....)))((((....((.(((....(((((((...........))))).))...(((.....-.)))))).))...))))...........((((((.....)))))).----- ( -12.80) >Banth.0 3158000 114 - 5227293/200-320 ACUGCUCCUAGGCGAGUUCAGGAUGUCAUUUGACUAUGUCACACCAACCCAUAGCUCUCUGUACAAAU-ACAUGACCUUACUACUCCUAUUCAACGCAUUUUAAUAUUUAGAAUA----- ..(((...((((.((((..(((.........(((((((...........))))).))((((((....)-))).))))).))).).))))......))).................----- ( -13.80) >consensus ACCACUCCCAGGCGAGUUCAAAAUGUCAUUUGACUACGUCACACCAACCCACAGCUCUCUGUACAAAU_ACAUGACCUUACUACUCCCAUUCAACGCAUUUUAAUAUUUAGAAUA_____ ..(((...((((.((((..(((((((.(((((((((((...........)))))........))))))...))))))).))).).))))......)))...................... (-21.14 = -16.07 + -5.06)

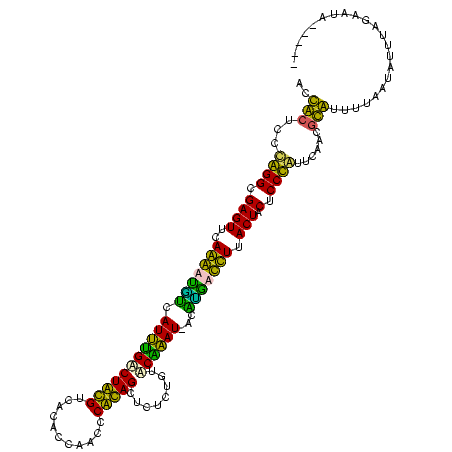
| Location | 3,496,180 – 3,496,282 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.05 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -24.51 |
| Energy contribution | -21.33 |
| Covariance contribution | -3.19 |
| Combinations/Pair | 1.57 |
| Mean z-score | 0.06 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 3496180 102 + 4222334/200-320 ------------------AAAACACAAUGAACGGGAGCAGUAAAAGAUCGGCAUUGGCAAAGAGAGCUGCGGGUAGGUGAGACGCAGACCAAUGAACCUUUGAACUCGCCCGGGAGUGGA ------------------....(((......((((.(.(((.((((.....((((((.........(((((...........))))).))))))...))))..)))).))))...))).. ( -25.40) >Bsubt.0 3490312 120 + 4214630/200-320 UGAGUUUUUCAGAAAACUAAAACACAUAGAUUGGGAGCAGUAAAAGAAAGUGAUCAGAACAGAGAGCUGCGGGGUGGUGCGACGCAGCCUGAUUCUUCUUUGAACUCGCCCGGGAGUGGU ..((((((....))))))....(((.....(((((.(.(((.((((((.(.((((((........((((((..(.....)..)))))))))))))))))))..)))).)))))..))).. ( -32.70) >Bcere.0 3070925 114 - 5224283/200-320 -----UAUUCUAAAUAUUAAAAUGUGUUGAAUAGGAGUAGUAAGGUCGUAU-AUUUGUACAGAGAGCUAUGGGUUGGUGUGACAUAGUCAAAUGACAUCCUGAACUCGCCUAGGAGCAGU -----...................((((...((((.(.(((.(((..((.(-(((((........((((((...........))))))))))))))..)))..)))).))))..)))).. ( -21.10) >Banth.0 3158000 114 - 5227293/200-320 -----UAUUCUAAAUAUUAAAAUGCGUUGAAUAGGAGUAGUAAGGUCAUGU-AUUUGUACAGAGAGCUAUGGGUUGGUGUGACAUAGUCAAAUGACAUCCUGAACUCGCCUAGGAGCAGU -----.................(((......((((.(.(((.(((..((((-(((((........((((((...........))))))))))).)))))))..)))).))))...))).. ( -20.60) >consensus _____UAUUCUAAAUAUUAAAACACAUUGAAUAGGAGCAGUAAAAGAAUGU_AUUUGUACAGAGAGCUACGGGUUGGUGUGACACAGUCAAAUGACACCCUGAACUCGCCCAGGAGCAGU ......................(((......((((.(.(((.((((..(((((((((........((((((...........)))))))))))))))))))..)))).))))...))).. (-24.51 = -21.33 + -3.19)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:26:10 2006