
| Sequence ID | Bcere.0 |
|---|---|
| Location | 5,077,163 – 5,077,283 |
| Length | 120 |
| Max. P | 0.999958 |

| Location | 5,077,163 – 5,077,276 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.19 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -13.65 |
| Energy contribution | -14.65 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 113 - 5224283/0-120 AUAGUUCUCUCAAAACUGAACAAAAC-GAAACACGGAAACUUAUAUUGAUGAACAGCGUU------CAUCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACAGCUAC ...((((..........)))).....-......(((((......((((((((((...)))------))))))).((((.......))))(((.......)))...))))).......... ( -24.20) >Banth.0 5080179 113 - 5227293/0-120 AUAGUUCUCUCAAAACUGAACAAAAC-GAAACACGGAAACUUAUAUUGAUGAACAGCGUU------CAUCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUAC ...((((..........)))).....-.......(((.......((((((((((...)))------)))))))(((((.......)))))...)))(((((............))))).. ( -27.20) >Blich.0 3121352 120 + 4222334/0-120 AAUGAUCCUUCAAAACUAAACAAGAUCCAAACGCACCUGUCUAUAAGACCGCAAGGUCUUAUAUUCCGUAUAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUAC ...((((................))))......((((....(((((((((....)))))))))............((((.....))))))))....(((((............))))).. ( -29.99) >Bsubt.0 3176052 119 + 4214630/0-120 AAUGAUCCUUCAAAACUAAACAAGACAGGGAACGUUCUGUUUAUAAGACCUUG-GGUCUUAUAUUCCGUAUAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUAC ...((((((((....((((....(((((((....)))))))(((((((((...-)))))))))...............))))....)))).)))).(((((............))))).. ( -29.80) >consensus AAAGAUCCCUCAAAACUAAACAAAAC_GAAACACGGAAACUUAUAAGAACGAACAGCCUU______CAUAAAAUAUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUAC ...((((((((....(((................(....).(((((((((....)))))))))................)))....)))).)))).(((((............))))).. (-13.65 = -14.65 + 1.00)


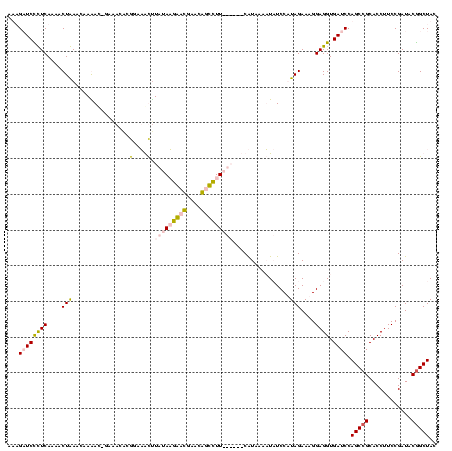
| Location | 5,077,163 – 5,077,276 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.19 |
| Mean single sequence MFE | -40.95 |
| Consensus MFE | -23.99 |
| Energy contribution | -26.80 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.79 |
| Structure conservation index | 0.59 |
| SVM decision value | 4.88 |
| SVM RNA-class probability | 0.999958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 113 - 5224283/0-120 GUAGCUGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGAUG------AACGCUGUUCAUCAAUAUAAGUUUCCGUGUUUC-GUUUUGUUCAGUUUUGAGAGAACUAU ..(((((((((....)))))))))(((.(((((((.......)))).(((((((------(((...))))))))))..........))).)))-(((((.(((......))))))))... ( -38.40) >Banth.0 5080179 113 - 5227293/0-120 GUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGAUG------AACGCUGUUCAUCAAUAUAAGUUUCCGUGUUUC-GUUUUGUUCAGUUUUGAGAGAACUAU ..(((((((((....)))))))))(((.(((((((.......)))).(((((((------(((...))))))))))..........))).)))-(((((.(((......))))))))... ( -40.60) >Blich.0 3121352 120 + 4222334/0-120 GUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUACGGAAUAUAAGACCUUGCGGUCUUAUAGACAGGUGCGUUUGGAUCUUGUUUAGUUUUGAAGGAUCAUU .((((((((((....))))))))))((((...((((.....)))).......((((((.(((((((....)))))))((((((((..(.....)..))))))))))))))...))))... ( -43.40) >Bsubt.0 3176052 119 + 4214630/0-120 GUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUACGGAAUAUAAGACC-CAAGGUCUUAUAAACAGAACGUUCCCUGUCUUGUUUAGUUUUGAAGGAUCAUU .((((((((((....))))))))))((((...((((.....)))).......((((((.(((((((-...)))))))((((((..(((.....)))..))))))))))))...))))... ( -41.40) >consensus GUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAGAAUAGACG______AACACCGUUCAGCAAUAUAAACAGCCGUGUUUC_GUCUUGUUCAGUUUUGAAAGAACAAU .((((((((((....))))))))))((((..((((.......))))...........(((((((((....)))))))))..................................))))... (-23.99 = -26.80 + 2.81)

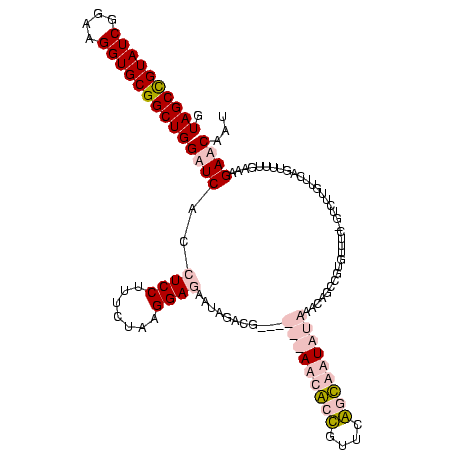

| Location | 5,077,163 – 5,077,277 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -43.92 |
| Consensus MFE | -31.89 |
| Energy contribution | -33.70 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.93 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 114 - 5224283/40-160 CCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCUGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGAUG------AACGCUGUUCAUCAAUAUAA (((.....))).((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))....(((((((------(((...)))))))))).... ( -40.40) >Banth.0 5080179 114 - 5227293/40-160 CCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGAUG------AACGCUGUUCAUCAAUAUAA (((.....))).((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))....(((((((------(((...)))))))))).... ( -42.60) >Blich.0 3121352 120 + 4222334/40-160 CCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUACGGAAUAUAAGACCUUGCGGUCUUAUAG (((..(((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....)))).......)))..(((((((((....))))))))). ( -47.10) >Bsubt.0 3176052 119 + 4214630/40-160 CCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUACGGAAUAUAAGACC-CAAGGUCUUAUAA (((..(((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....)))).......)))..(((((((((-...))))))))). ( -45.60) >consensus CCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAGAAUAGACG______AACACCGUUCAGCAAUAUAA .....(((((..(..((((((.......))))))........(((((((((....))))))))))..)))))(((.......)))............(((((((((....))))))))). (-31.89 = -33.70 + 1.81)

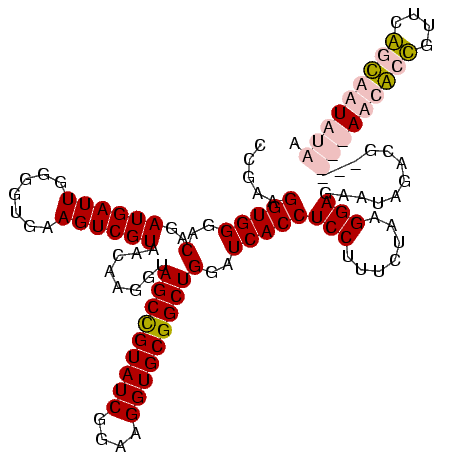
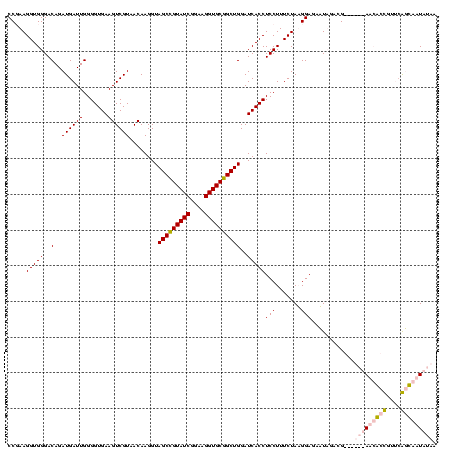
| Location | 5,077,163 – 5,077,283 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -47.83 |
| Consensus MFE | -46.89 |
| Energy contribution | -47.70 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 120 - 5224283/80-200 ACACCCGAAGUCGGUGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCUGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCU ...((((.....((((((((..((.....)).)))..))))).....))))......((..(((((((.((....))....((((((((((....))))))))))..))))).))..)). ( -45.00) >Banth.0 5080179 120 - 5227293/80-200 ACACCCGAAGUCGGUGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCU ...((((.....((((((((..((.....)).)))..))))).....))))......((..(((((((.((....))....((((((((((....))))))))))..))))).))..)). ( -47.20) >Blich.0 3121352 119 + 4222334/80-200 ACACCCGAAGUCGGUGAGGUAACCUUUU-GGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCU ...((((...((((((.(((..((....-)).)))...))))))...))))......((..(((((((.((....))....((((((((((....))))))))))..))))).))..)). ( -49.70) >Bsubt.0 3176052 120 + 4214630/80-200 ACACCCGAAGUCGGUGAGGUAACCUUUUAGGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCU ...((((...((((((.(((..((.....)).)))...))))))...))))......((..(((((((.((....))....((((((((((....))))))))))..))))).))..)). ( -49.40) >consensus ACACCCGAAGUCGGUGAGGUAACCUUUUUGGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCU ...((((...((((((((((..((.....)).)))..)))))))...))))......((..(((((((.((....))....((((((((((....))))))))))..))))).))..)). (-46.89 = -47.70 + 0.81)

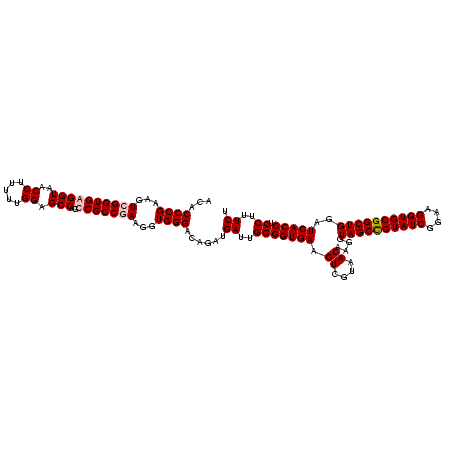
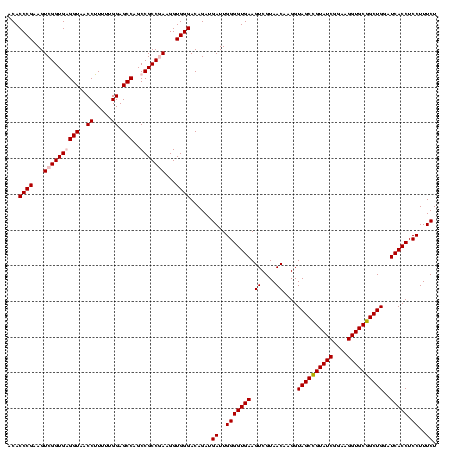
| Location | 5,077,163 – 5,077,283 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -45.40 |
| Consensus MFE | -46.90 |
| Energy contribution | -45.40 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.51 |
| Structure conservation index | 1.03 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 120 - 5224283/200-320 UACAAACUCUCGUGGUGUGACGGGCGGUGUGUACAAGGCCCGGGAACGUAUUCACCGCGGCAUGCUGAUCCGCGAUUACUAGCGAUUCCAGCUUCAUGUAGGCGAGUUGCAGCCUACAAU .........((((((((..((((((..((....))..))))(....)))...))))))))...(((((..(((........)))..).))))....(((((((........))))))).. ( -44.80) >Banth.0 5080179 120 - 5227293/200-320 UACAAACUCUCGUGGUGUGACGGGCGGUGUGUACAAGGCCCGGGAACGUAUUCACCGCGGCAUGCUGAUCCGCGAUUACUAGCGAUUCCAGCUUCAUGUAGGCGAGUUGCAGCCUACAAU .........((((((((..((((((..((....))..))))(....)))...))))))))...(((((..(((........)))..).))))....(((((((........))))))).. ( -44.80) >Blich.0 3121352 120 + 4222334/200-320 UACAAACUCUCGUGGUGUGACGGGCGGUGUGUACAAGGCCCGGGAACGUAUUCACCGCGGCAUGCUGAUCCGCGAUUACUAGCGAUUCCAGCUUCACGCAGUCGAGUUGCAGACUGCGAU .........((((((((..((((((..((....))..))))(....)))...))))))))...(((((..(((........)))..).))))....(((((((........))))))).. ( -46.00) >Bsubt.0 3176052 120 + 4214630/200-320 UACAAACUCUCGUGGUGUGACGGGCGGUGUGUACAAGGCCCGGGAACGUAUUCACCGCGGCAUGCUGAUCCGCGAUUACUAGCGAUUCCAGCUUCACGCAGUCGAGUUGCAGACUGCGAU .........((((((((..((((((..((....))..))))(....)))...))))))))...(((((..(((........)))..).))))....(((((((........))))))).. ( -46.00) >consensus UACAAACUCUCGUGGUGUGACGGGCGGUGUGUACAAGGCCCGGGAACGUAUUCACCGCGGCAUGCUGAUCCGCGAUUACUAGCGAUUCCAGCUUCACGCAGGCGAGUUGCAGACUACAAU .........((((((((..((((((..((....))..))))(....)))...))))))))...(((((..(((........)))..).))))....(((((((........))))))).. (-46.90 = -45.40 + -1.50)

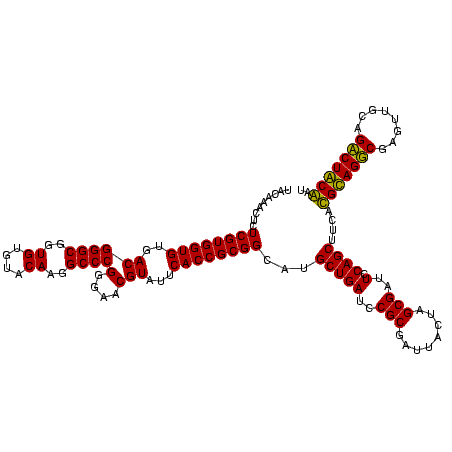
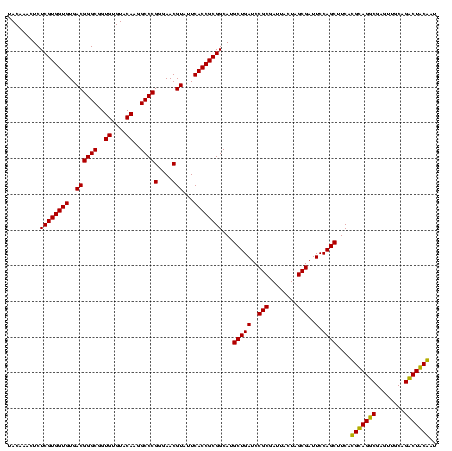
| Location | 5,077,163 – 5,077,283 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -39.29 |
| Energy contribution | -36.10 |
| Covariance contribution | -3.19 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.11 |
| Structure conservation index | 1.06 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 120 - 5224283/280-400 CACACGUGCUACAAUGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCGCU .......((......(((((((.....(((..((...(((....)))...))....))).....)))))))((((((.....(((((((........)))))))...))))))....)). ( -35.30) >Banth.0 5080179 120 - 5227293/280-400 CACACGUGCUACAAUGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCGCU .......((......(((((((.....(((..((...(((....)))...))....))).....)))))))((((((.....(((((((........)))))))...))))))....)). ( -35.30) >Blich.0 3121352 120 + 4222334/280-400 CACACGUGCUACAAUGGGCAGAACAAAGGGCAGCGAAGCCGCGAGGCUAAGCCAAUCCCACAAAUCUGUUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGCUGGAAUCGCU .......((......(((((((.....(((..((..((((....))))..))....))).....)))))))((((((.....(((((((........)))))))...))))))....)). ( -40.80) >Bsubt.0 3176052 119 + 4214630/280-400 CACACGUGCUACAAUGGACAGAACAAAGGGCAGCGAA-CCGCGAGGUUAAGCCAAUCCCACAAAUCUGUUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGCUGGAAUCGCU .......((......(((((((.....(((..((.((-((....))))..))....))).....)))))))((((((.....(((((((........)))))))...))))))....)). ( -37.40) >consensus CACACGUGCUACAAUGGACAGAACAAAGAGCAGCAAAACCGCGAGGUGAAGCCAAUCCCACAAAACCGUUCUCAGUUCGGAUCGCAGGCUGCAACUCGACUACAUGAAGCUGGAAUCGCU .......((......(((((((.....(((..((...(((....)))...))....))).....)))))))((((((.....(((((((........)))))))...))))))....)). (-39.29 = -36.10 + -3.19)

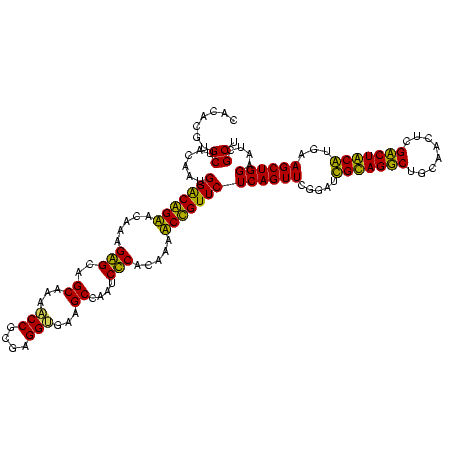

| Location | 5,077,163 – 5,077,283 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -48.72 |
| Consensus MFE | -49.30 |
| Energy contribution | -48.17 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.44 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 120 - 5224283/680-800 CUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUG ...((..(((((((((....)))))))))..))................((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).. ( -48.20) >Banth.0 5080179 120 - 5227293/680-800 CUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUG ...((..(((((((((....)))))))))..))................((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).. ( -48.20) >Blich.0 3121352 120 + 4222334/680-800 CUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGCAGCAAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUG ...((..(((((((((....)))))))))..))...((....)).....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).. ( -47.30) >Bsubt.0 3176052 120 + 4214630/680-800 CUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUGAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUG ...((..(((((((((....)))))))))..))...((....)).....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).. ( -51.20) >consensus CUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUG ...((..(((((((((....)))))))))..))...((....)).....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).. (-49.30 = -48.17 + -1.12)

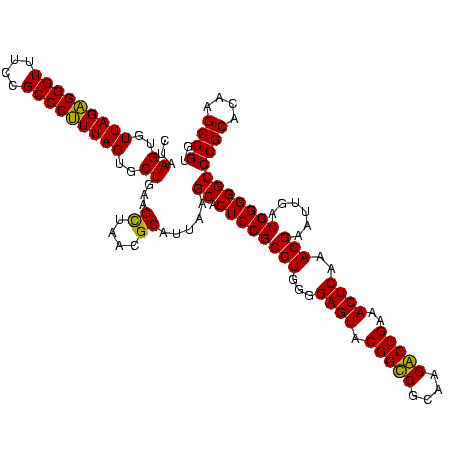
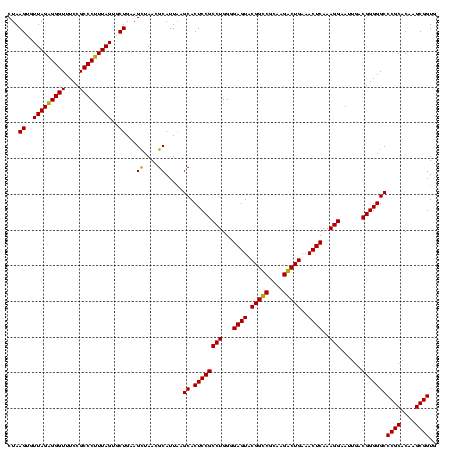
| Location | 5,077,163 – 5,077,283 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -34.59 |
| Consensus MFE | -31.58 |
| Energy contribution | -32.40 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 120 - 5224283/760-880 UAACUUCAGCACUAAAGGGCGGAAACCCUCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCGCCUCAGUGUCAGUUAC (((((...(((((..(((((((((.......................((....))((((((.....((((((.....))))))........)))))))))))).))).))))).))))). ( -33.42) >Banth.0 5080179 120 - 5227293/760-880 UAACUUCAGCACUAAAGGGCGGAAACCCUCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCGCCUCAGUGUCAGUUAC (((((...(((((..(((((((((.......................((....))((((((.....((((((.....))))))........)))))))))))).))).))))).))))). ( -33.42) >Blich.0 3121352 120 + 4222334/760-880 UUGCUGCAGCACUAAAGGGCGGAAACCCUCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUCGCUCCCCACGCUUUCGCGCCUCAGCGUCAGUUAC ..((((..((.....((((.(....).))))........((......((....))((((((.....((((((.....))))))........))))))....))))..))))......... ( -32.92) >Bsubt.0 3176052 120 + 4214630/760-880 UAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUCGCUCCCCACGCUUUCGCUCCUCAGCGUCAGUUAC ((((((..((.((.((((((((((.......................((....))((((((.....((((((.....))))))........)))))))))))))))).)).)))))))). ( -38.62) >consensus UAACUGCAGCACUAAAGGGCGGAAACCCUCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUCGCUCCCCACGCUUUCGCGCCUCAGCGUCAGUUAC ((((((..(((((..(((((((((.......................((....))((((((.....((((((.....))))))........)))))))))))).))).))))))))))). (-31.58 = -32.40 + 0.81)

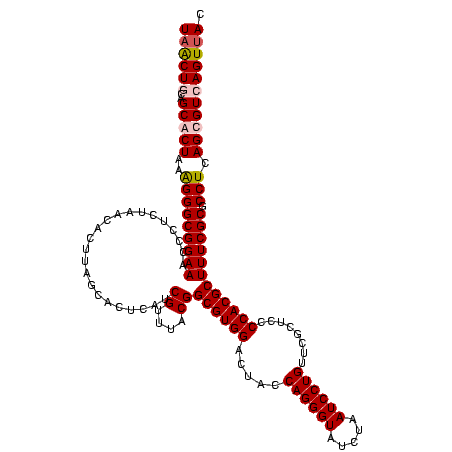
| Location | 5,077,163 – 5,077,283 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -40.60 |
| Energy contribution | -40.48 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 120 - 5224283/760-880 GUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUA .(((((..((((.((((((....))(((((..((...((((.........))))))..))))))))(....)..).))))...((..(((((((((....)))))))))..))..))))) ( -41.60) >Banth.0 5080179 120 - 5227293/760-880 GUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUA .(((((..((((.((((((....))(((((..((...((((.........))))))..))))))))(....)..).))))...((..(((((((((....)))))))))..))..))))) ( -41.60) >Blich.0 3121352 120 + 4222334/760-880 GUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGCAGCAA .........((((.(((((....((((((...((...((((.........))))...))))))))((....))....))))).((..(((((((((....)))))))))..)).)))).. ( -45.20) >Bsubt.0 3176052 120 + 4214630/760-880 GUAACUGACGCUGAGGAGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUA ...(((.(((((....))))...((((((...((...((((.........))))...)))))))).........).)))(((.((..(((((((((....)))))))))..))..))).. ( -43.20) >consensus GUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGCUA .........((((.(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).((..(((((((((....)))))))))..)).)))).. (-40.60 = -40.48 + -0.12)

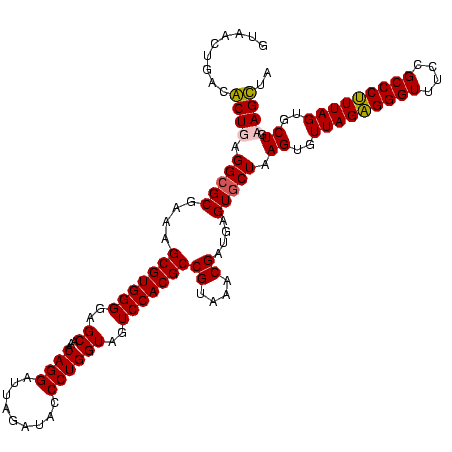
| Location | 5,077,163 – 5,077,282 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.82 |
| Mean single sequence MFE | -40.12 |
| Consensus MFE | -34.85 |
| Energy contribution | -34.35 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 119 - 5224283/840-960 GAAAGUGGAAUUCCAUG-UGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAG ....(((...(((((((-(.(.(((......)))....).))))))))...)))(((((((....))((((....)))).........)))))..((((....))))............. ( -38.20) >Banth.0 5080179 119 - 5227293/840-960 GAAAGUGGAAUUCCAUG-UGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAG ....(((...(((((((-(.(.(((......)))....).))))))))...)))(((((((....))((((....)))).........)))))..((((....))))............. ( -38.20) >Blich.0 3121352 119 + 4222334/840-960 GAGAGUGGAAUUCCACG-UGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCGAACAG (((((((...(((((((-(.(.(((......)))....).))))))))...)))..((.((....)).))))))..((.(((..((.((((....))((....)))).))..))).)).. ( -42.20) >Bsubt.0 3176052 120 + 4214630/840-960 GAGAGUGGAAUUCCACGUUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGAGCGAAAGCGUGGGGAGCGAACAG (((((((...(((((((((...(((......))).....)))))))))...)))..((.((....)).))))))..((.(((..((.(((((..........))))).))..))).)).. ( -41.90) >consensus GAAAGUGGAAUUCCACG_UGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAG ....(((...(((((((.....(((......))).......)))))))...)))(((((((....))((((....)))).........)))))..((((....))))............. (-34.85 = -34.35 + -0.50)


| Location | 5,077,163 – 5,077,282 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.23 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -33.97 |
| Energy contribution | -32.73 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 119 - 5224283/880-1000 GGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUG-UGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCU .(((..((...(....)...))..))).....(((((.......(((...(((((((-(.(.(((......)))....).))))))))...))).(((.((....)).)))))))).... ( -34.20) >Banth.0 5080179 119 - 5227293/880-1000 GGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUG-UGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCU .(((..((...(....)...))..))).....(((((.......(((...(((((((-(.(.(((......)))....).))))))))...))).(((.((....)).)))))))).... ( -34.20) >Blich.0 3121352 119 + 4222334/880-1000 GGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACG-UGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCU ((((((((.......(((((...((((.....(....)....))))(...(((((((-(.(.(((......)))....).))))))))...).))))).((....))))))))))..... ( -37.50) >Bsubt.0 3176052 120 + 4214630/880-1000 GGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCU ((((((((.......(((((...((((.....(....)....))))(...(((((((((...(((......))).....)))))))))...).))))).((....))))))))))..... ( -38.40) >consensus GGAGGGUCAUUGGAAACUGGGAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCACG_UGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCU .(((..((...(....)...))..)))......(((...((((((((...(((((((.....(((......))).......)))))))...)))..((.((....)).)))))))..))) (-33.97 = -32.73 + -1.25)

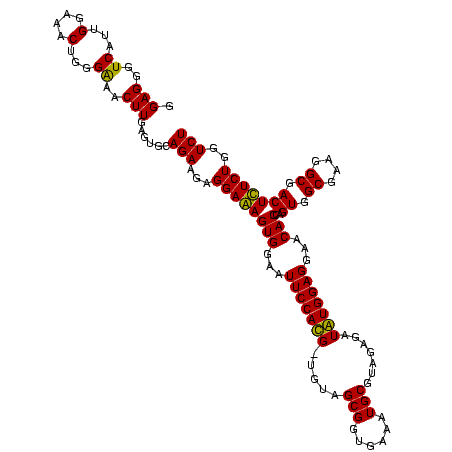
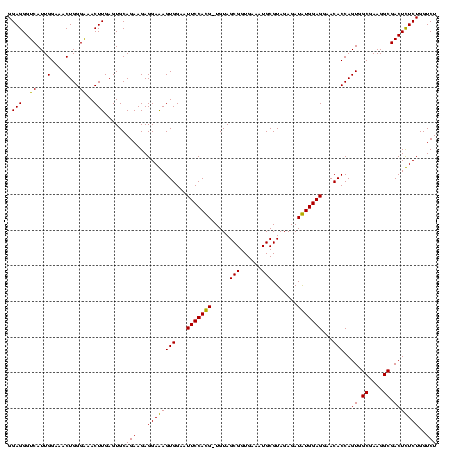
| Location | 5,077,163 – 5,077,282 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.23 |
| Mean single sequence MFE | -24.41 |
| Consensus MFE | -22.20 |
| Energy contribution | -21.95 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 119 - 5224283/920-1040 CUCUACGCAUUUCACCGCUACA-CAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAGAAACC .........((((...((....-...((((.......))))......))....((((((....(((......)))..(((((((.....)))))))..........))))))..)))).. ( -24.02) >Banth.0 5080179 119 - 5227293/920-1040 CUCUACGCAUUUCACCGCUACA-CAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAGAAACC .........((((...((....-...((((.......))))......))....((((((....(((......)))..(((((((.....)))))))..........))))))..)))).. ( -24.02) >Blich.0 3121352 119 + 4222334/920-1040 CUCUACGCAUUUCACCGCUACA-CGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUUCCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAGAAACC ......((........))....-.((((....))))....................(((...(((((....(((...(((((((.....)))))))...)))....)))))...)))... ( -24.80) >Bsubt.0 3176052 120 + 4214630/920-1040 CUCUACGCAUUUCACCGCUACAACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUUCCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAGAAACC ......((........))......((((....))))....................(((...(((((....(((...(((((((.....)))))))...)))....)))))...)))... ( -24.80) >consensus CUCUACGCAUUUCACCGCUACA_CAUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUCCCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAGAAACC ......((........))......((((....))))..............((.(((((.....(((......)))..(((((((.....)))))))...........))))).))..... (-22.20 = -21.95 + -0.25)

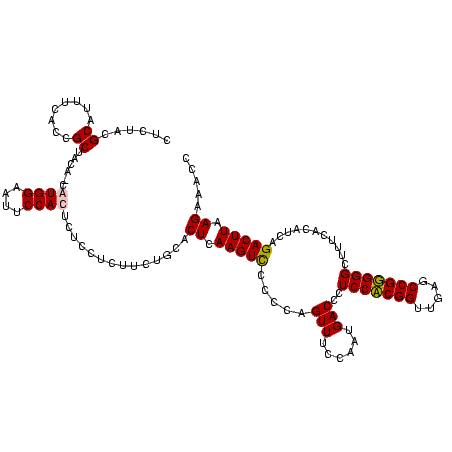
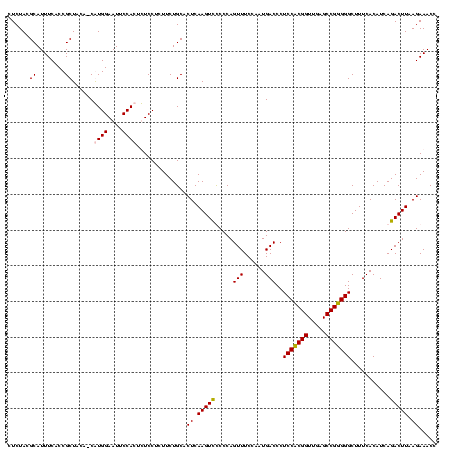
| Location | 5,077,163 – 5,077,283 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -43.00 |
| Consensus MFE | -44.92 |
| Energy contribution | -43.18 |
| Covariance contribution | -1.75 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.70 |
| Structure conservation index | 1.04 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 120 - 5224283/1080-1200 GUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCA .......((((..((((((......((((((((((((.....)))))))).))))......))))))))))..(((((......(((((((((.((....)))))...))))))))))). ( -43.70) >Banth.0 5080179 120 - 5227293/1080-1200 GUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCA .......((((..((((((......((((((((((((.....)))))))).))))......))))))))))..(((((......(((((((((.((....)))))...))))))))))). ( -43.70) >Blich.0 3121352 120 + 4222334/1080-1200 GUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCA .......((((..((((((......((((..((((((.....))))))...))))......))))))))))..(((((......(((((((((.((....)))))...))))))))))). ( -40.20) >Bsubt.0 3176052 120 + 4214630/1080-1200 GUAAAGCUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAACAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCA .......((((..((((((......((((((((((((.....)))))))).))))......))))))))))..(((((......(((((((((.((....)))))...))))))))))). ( -44.40) >consensus GUAAAACUCUGUUGUUAGGGAAGAACAAGUACCAGUCGAAUAAGCCGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCA .......((((..((((((......((((((((((((.....)))))))).))))......))))))))))..(((((......(((((((((.((....)))))...))))))))))). (-44.92 = -43.18 + -1.75)

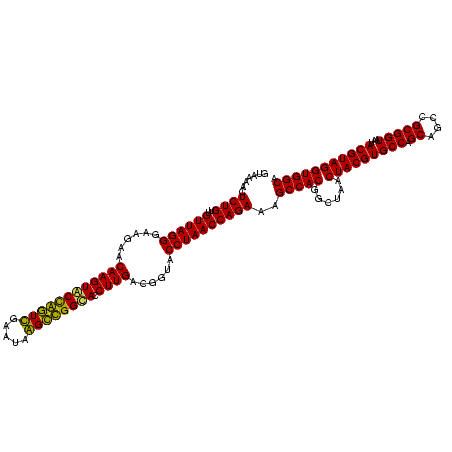

| Location | 5,077,163 – 5,077,283 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.19 |
| Mean single sequence MFE | -41.28 |
| Consensus MFE | -41.01 |
| Energy contribution | -39.33 |
| Covariance contribution | -1.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 120 - 5224283/1120-1240 GACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGG ..((......((.(((....))).))..(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..))))))).)). ( -43.20) >Banth.0 5080179 120 - 5227293/1120-1240 GACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGG ..((......((.(((....))).))..(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..))))))).)). ( -43.20) >Blich.0 3121352 120 + 4222334/1120-1240 GACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGCACCUUGACGGUACCUAACCAGAAAGCCACGG ..((..((..((.(((....))).))..((((...(((((((....(((((....))))).....((((..((((((.....))))))...))))))))).)).)))).....))..)). ( -37.00) >Bsubt.0 3176052 120 + 4214630/1120-1240 GACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAACAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGG ((((((((..((.(((....))).))..((((....)))).....))))))))((((((......((((((((((((.....)))))))).))))......))))))............. ( -41.70) >consensus GACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCAGUCGAAUAAGCCGGCACCUUGACGGUACCUAACCAGAAAGCCACGG ..((......((.(((....))).))..(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..))))))).)). (-41.01 = -39.33 + -1.69)

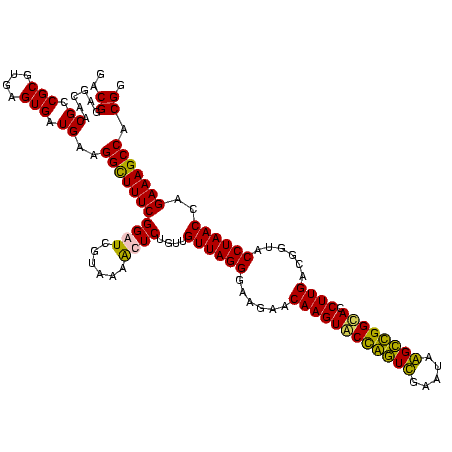
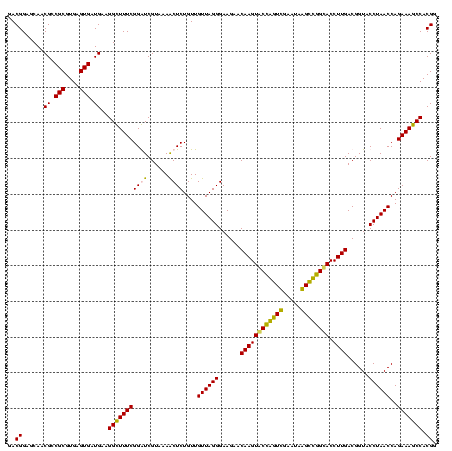
| Location | 5,077,163 – 5,077,283 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -48.35 |
| Consensus MFE | -48.24 |
| Energy contribution | -48.05 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.29 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 120 - 5224283/1240-1360 UCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCU ....((..((.((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...))..(((....)))))..))(((....))). ( -48.40) >Banth.0 5080179 120 - 5227293/1240-1360 UCACCAAGGCAAUGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCU ....((..((..((.(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))))).))....(((....)))))..))(((....))). ( -48.10) >Blich.0 3121352 120 + 4222334/1240-1360 UCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCU ....((..((.((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...))..(((....)))))..))(((....))). ( -48.50) >Bsubt.0 3176052 120 + 4214630/1240-1360 UCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCU ....((..((.((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...))..(((....)))))..))(((....))). ( -48.40) >consensus UCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCU ....((..((.((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...))..(((....)))))..))(((....))). (-48.24 = -48.05 + -0.19)

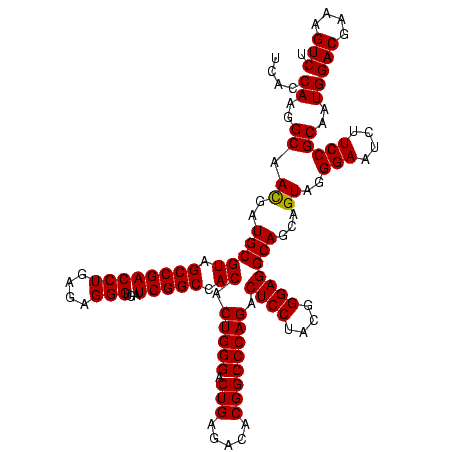
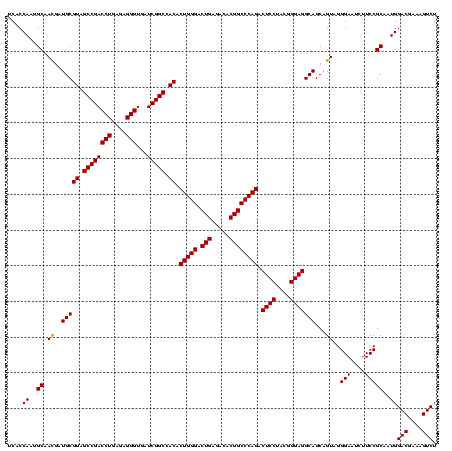
| Location | 5,077,163 – 5,077,283 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -48.83 |
| Consensus MFE | -47.08 |
| Energy contribution | -47.57 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 120 - 5224283/1280-1400 CCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAU ...........(((((((((((.((......((((((((((((.........))))))))))))...((((..((((((((.......)))))))).))))...)).))))).)))))). ( -50.20) >Banth.0 5080179 120 - 5227293/1280-1400 CCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCAUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAU ...........(((((((((((.........((((((((((((.........))))))))))))..((((((.((((((((.......))))))))...)).)))).))))).)))))). ( -50.30) >Blich.0 3121352 120 + 4222334/1280-1400 CCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAU .....(((..(((((......))))).(((.((((((((((((.........))))))))))))...((.((.((((((((.......))))))))...((....)))).)))))))).. ( -47.40) >Bsubt.0 3176052 120 + 4214630/1280-1400 CCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAU .....(((..(((((......))))).(((.((((((((((((.........))))))))))))...((.((.((((((((.......))))))))...((....)))).)))))))).. ( -47.40) >consensus CCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAU ...........(((((((((((.((......((((((((((((.........))))))))))))......((.((((((((.......))))))))...))...)).))))).)))))). (-47.08 = -47.57 + 0.50)

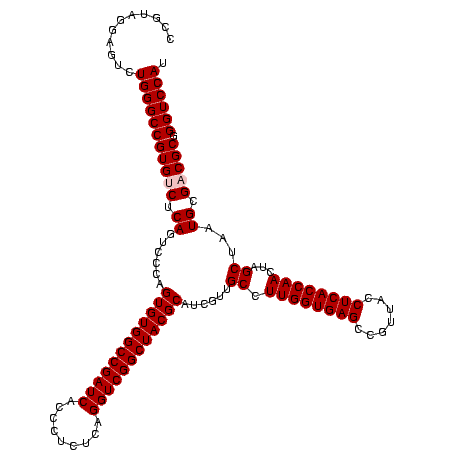
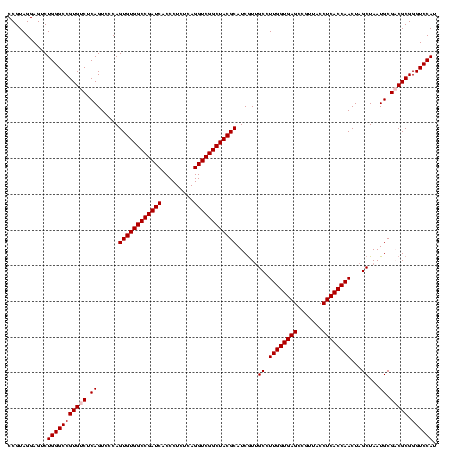
| Location | 5,077,163 – 5,077,281 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.18 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -32.38 |
| Energy contribution | -32.25 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 118 - 5224283/1320-1440 AUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCCGAA-GCCGCCUUUCA-AUUUCGAAC ...........(((.(((((.(((.((((.((.((((((((.......))))))))...))....)))).)))(((.(((......)))...)))..)-)))))))....-......... ( -35.50) >Banth.0 5080179 118 - 5227293/1320-1440 AUCACCCUCUCAGGUCGGCUACGCAUCAUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGGCAGCCGAA-GCCACCUUUCA-AUUUCGAAC ...........((((.((((.(((((....((.((((((((.......))))))))...))..)))))...(((.(((((......)))).).))).)-)))))))....-......... ( -36.20) >Blich.0 3121352 120 + 4222334/1320-1440 AUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGGUAGCUGAAAGCCACCUUUUAUGAUUGGAAC ....(((.....(((.(((.(((...))).)))((((((((.......))))))))...((....)))))..)))((((..((((((.(((.((.....)).))).))))))..)))).. ( -37.00) >Bsubt.0 3176052 118 + 4214630/1320-1440 AUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGGUAGCCGAA-GCCACCUUUUAUGUUUG-AAC .(((..(....((((.((((.(((((....((.((((((((.......))))))))...))..)))))...(((.(((((......)))).).))).)-))))))).....)..))-).. ( -37.20) >consensus AUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGGCAGCCGAA_GCCACCUUUCA_AUUUCGAAC ...........((((.(((..(((((....((.((((((((.......))))))))...))..)))))...(((.(((((......)))).).)))...))))))).............. (-32.38 = -32.25 + -0.12)

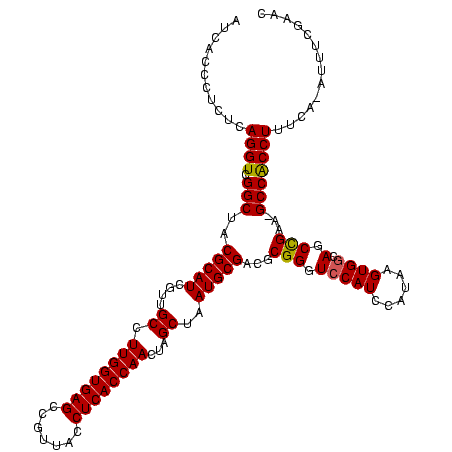
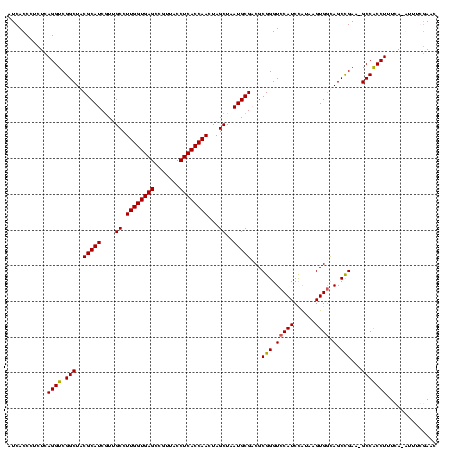
| Location | 5,077,163 – 5,077,281 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.18 |
| Mean single sequence MFE | -45.40 |
| Consensus MFE | -38.90 |
| Energy contribution | -38.27 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 118 - 5224283/1320-1440 GUUCGAAAU-UGAAAGGCGGC-UUCGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAU (..(.....-.....((((((-....)))))).((((.((.(((...(((((((.....(((..((((((((.......)))))))))))..)))))))...))).)))))).)..)... ( -44.90) >Banth.0 5080179 118 - 5227293/1320-1440 GUUCGAAAU-UGAAAGGUGGC-UUCGGCUGCCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAAUGAUGCGUAGCCGACCUGAGAGGGUGAU (((((....-(((..((..((-....))..))..)))....)))))..((..((((((((((..((((((((.......)))))))))))..)))))))..))..((((....))))... ( -44.70) >Blich.0 3121352 120 + 4222334/1320-1440 GUUCCAAUCAUAAAAGGUGGCUUUCAGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAU ..((((.((.(((..((((((.....))))))..)))..))))))...((.(((((((.(((..((((((((.......)))))))))))...))))))).))..((((....))))... ( -45.30) >Bsubt.0 3176052 118 + 4214630/1320-1440 GUU-CAAACAUAAAAGGUGGC-UUCGGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAU (((-((....(((..((((((-....))))))..)))....)))))..((.(((((((.(((..((((((((.......)))))))))))...))))))).))..((((....))))... ( -46.70) >consensus GUUCCAAAC_UAAAAGGUGGC_UUCGGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAU ((((......(((..((((((.....))))))..))).....))))..((..((((((.(((..((((((((.......)))))))))))...))))))..))..((((....))))... (-38.90 = -38.27 + -0.63)

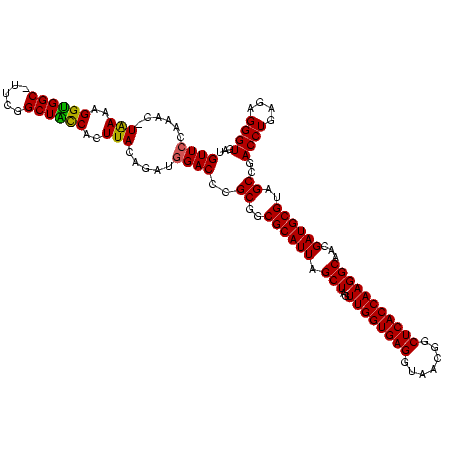
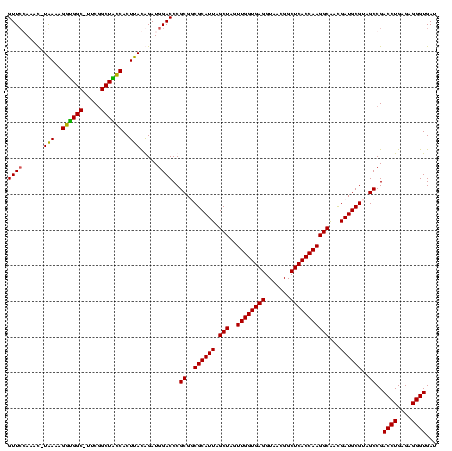
| Location | 5,077,163 – 5,077,281 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.66 |
| Mean single sequence MFE | -41.95 |
| Consensus MFE | -32.33 |
| Energy contribution | -33.08 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 118 - 5224283/1360-1480 GAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAU-UGAAAGGCGGC-UUCGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGC ....(((..(((..(((((......((((((((((....))))))))))-.....((((((-....))))))(((....(((((((....))).))))....)))))))).)))..))). ( -39.60) >Banth.0 5080179 118 - 5227293/1360-1480 GAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAU-UGAAAGGUGGC-UUCGGCUGCCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGC ....(((..(((..(((((......((((((((((....))))))))))-.....((..((-....))..))(((....(((((((....))).))))....)))))))).)))..))). ( -39.80) >Blich.0 3121352 120 + 4222334/1360-1480 GAAACCGGGGCUAAUACCGGAUGCUUGAUUGAACCGCAUGGUUCCAAUCAUAAAAGGUGGCUUUCAGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGC ....(((..(((..((((((..(((....((..((((..(((.(((.((.(((..((((((.....))))))..)))..)))))))).))))..))..)))...)))))).)))..))). ( -41.70) >Bsubt.0 3176052 118 + 4214630/1360-1480 GAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGCAUGGUU-CAAACAUAAAAGGUGGC-UUCGGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGC ....(((..(((..((((((.((((((..((..((((..((((-((....(((..((((((-....))))))..)))....)))))).))))..)).)))))).)))))).)))..))). ( -46.70) >consensus GAAACCGGGGCUAAUACCGGAUAACAGUUUGAACCGCAUGGUUCCAAAC_UAAAAGGUGGC_UUCGGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGC ....(((..(((....(((..((((((((((((((....))))))))))))....((((((.....)))))).....))..)))(((.((...((....))..)).)))..)))..))). (-32.33 = -33.08 + 0.75)

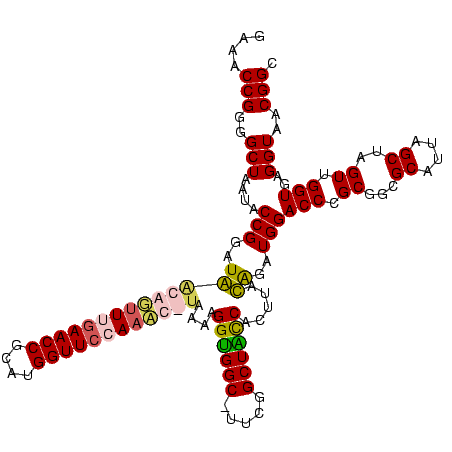
| Location | 5,077,163 – 5,077,281 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.11 |
| Mean single sequence MFE | -37.02 |
| Consensus MFE | -30.34 |
| Energy contribution | -31.15 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 118 - 5224283/1400-1520 CCAUAAGUGACAGCCGAA-GCCGCCUUUCA-AUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCAGGUUACCCACGUGU ((((((((((((.....(-(((((..(((.-.....)))....)))))).....)))))...((...((((.((((....)))).))))..))...)))))))(((.((.....)).))) ( -33.90) >Banth.0 5080179 118 - 5227293/1400-1520 CCAUAAGUGGCAGCCGAA-GCCACCUUUCA-AUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCAGGUUACCCACGUGU (((((((((((.......-)))))......-((((.(((((....))))).)))).......((...((((.((((....)))).))))..))....))))))(((.((.....)).))) ( -33.70) >Blich.0 3121352 120 + 4222334/1400-1520 CUGUAAGUGGUAGCUGAAAGCCACCUUUUAUGAUUGGAACCAUGCGGUUCAAUCAAGCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGU (((((((.((..((((((((....))))).((((((.((((....)))))))))))))..))((...((((.((((....)))).))))..))...)))))))(((.((.....)).))) ( -37.50) >Bsubt.0 3176052 118 + 4214630/1400-1520 CUGUAAGUGGUAGCCGAA-GCCACCUUUUAUGUUUG-AACCAUGCGGUUCAAACAACCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGU ......(((((((((...-(((........((((((-((((....)))))))))).......((...((((.((((....)))).))))..)).........))).))))).)))).... ( -43.00) >consensus CCAUAAGUGGCAGCCGAA_GCCACCUUUCA_AUUUCGAACCAUGCGGUUCAAAAAAUCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGU ......(((((((((....(((..............(((((....)))))............((...((((.((((....)))).))))..)).........))).))))).)))).... (-30.34 = -31.15 + 0.81)

| Location | 5,077,163 – 5,077,281 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.11 |
| Mean single sequence MFE | -52.12 |
| Consensus MFE | -44.90 |
| Energy contribution | -45.40 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -5.17 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 118 - 5224283/1400-1520 ACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAU-UGAAAGGCGGC-UUCGGCUGUCACUUAUGG ....(..((...))..)(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....))))))))))-.....((((((-....)))))).))))))) ( -51.70) >Banth.0 5080179 118 - 5227293/1400-1520 ACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAU-UGAAAGGUGGC-UUCGGCUGCCACUUAUGG ....(..((...))..)(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....))))))))))-.....((..((-....))..)).))))))) ( -51.90) >Blich.0 3121352 120 + 4222334/1400-1520 ACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAACCGCAUGGUUCCAAUCAUAAAAGGUGGCUUUCAGCUACCACUUACAG ....(..((...))..)(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))).)))))).....((((((.....)))))).))))))) ( -49.20) >Bsubt.0 3176052 118 + 4214630/1400-1520 ACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGCAUGGUU-CAAACAUAAAAGGUGGC-UUCGGCUACCACUUACAG ....(..((...))..)(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....))))-)))))).....((((((-....)))))).))))))) ( -55.70) >consensus ACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAGUUUGAACCGCAUGGUUCCAAAC_UAAAAGGUGGC_UUCGGCUACCACUUACAG ....(..((...))..)(((((((.((((..((.((((((....)))))).))...))))...((((((((((((....))))))))))))....((((((.....)))))).))))))) (-44.90 = -45.40 + 0.50)

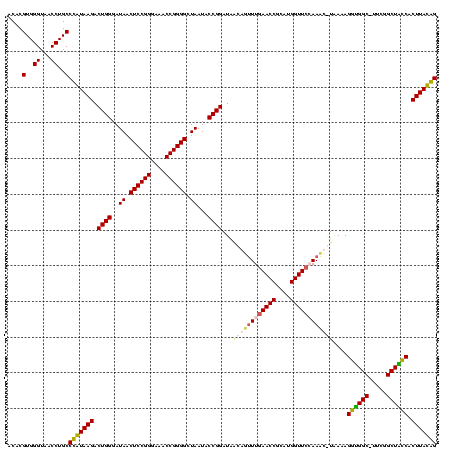
| Location | 5,077,163 – 5,077,283 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.24 |
| Mean single sequence MFE | -42.47 |
| Consensus MFE | -40.55 |
| Energy contribution | -40.30 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 5077163 120 - 5224283/1440-1560 AGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUG ...(((.((((....).))).))).((((..((((..((.((......)).))..))))......((((..((.((((((....)))))).))...)))).............))))... ( -41.60) >Banth.0 5080179 120 - 5227293/1440-1560 AGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUG ...(((.((((....).))).))).((((..((((..((.((......)).))..))))......((((..((.((((((....)))))).))...)))).............))))... ( -41.60) >Blich.0 3121352 119 + 4222334/1440-1560 GGAGCUUGCUCCC-UUAGGUCAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAACCGCAUG ((((....)))).-...........((((((((((..((.((......)).))..))))))(((.((((..((.((((((....)))))).))...))))...))).......))))... ( -42.90) >Bsubt.0 3176052 117 + 4214630/1440-1560 GGAGCUUGCUCC---GAUGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGCAUG ((((....))))---.......(((((..((((((..((.((......)).))..))))))..).((((..((.((((((....)))))).))...)))).............))))... ( -43.80) >consensus AGAGCUUGCUCCU_UGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAGUUUGAACCGCAUG ((((....))))..........(((((..((((((..((.((......)).))..))))))..).((((..((.((((((....)))))).))...)))).............))))... (-40.55 = -40.30 + -0.25)

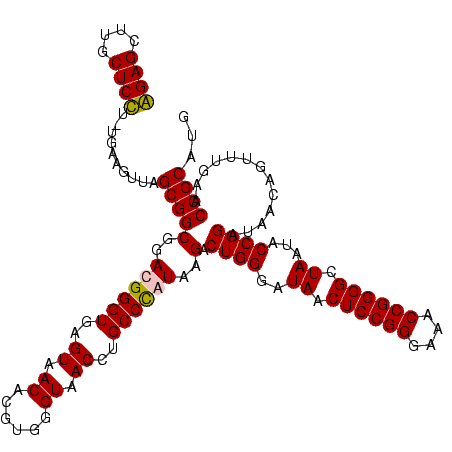
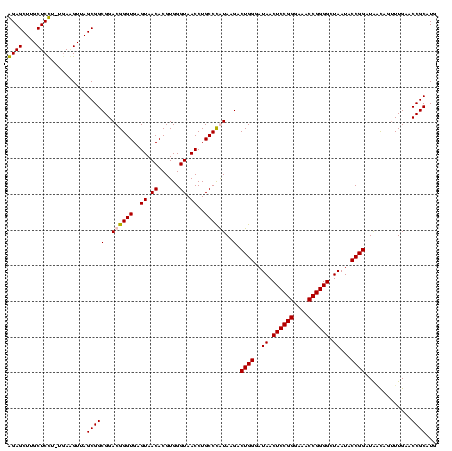
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:26:06 2006