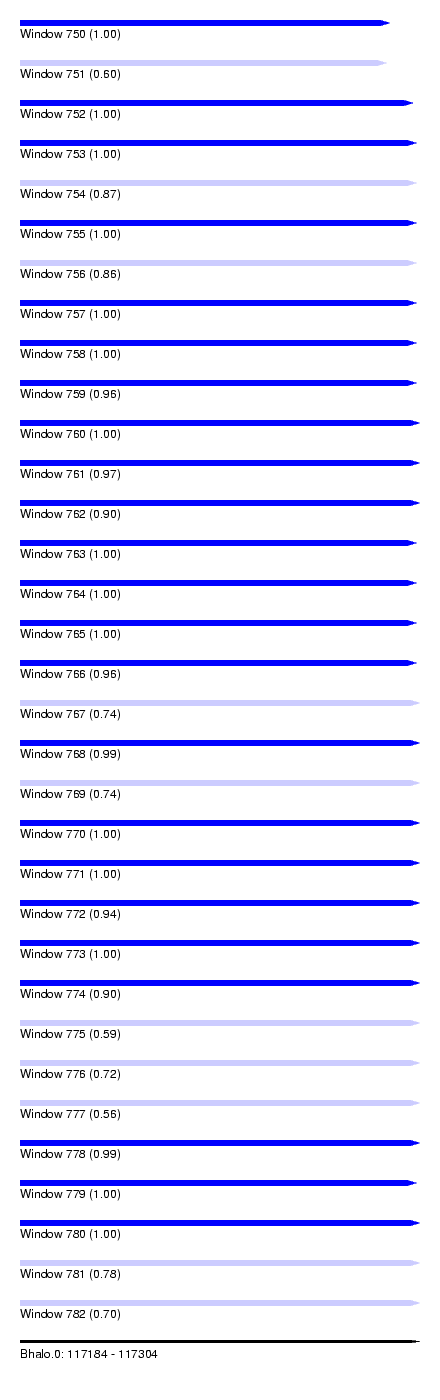
| Sequence ID | Bhalo.0 |
|---|---|
| Location | 117,184 – 117,304 |
| Length | 120 |
| Max. P | 0.999998 |

| Location | 117,184 – 117,295 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.11 |
| Mean single sequence MFE | -44.55 |
| Consensus MFE | -21.27 |
| Energy contribution | -20.90 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.46 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.48 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 111 + 4202352/160-280 CUUCAUUAUAUGAAGUGUCACAGGAAACAACAUCCUGUGAAAC-AUGAGCUUUAUC----AAACUUUUAU----GGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAA ((((((...))))))..((((((((.......))))))))...-..(((((..(((----((((((((..----.)))))))))))...)))))(((....((((.....)))).))).. ( -40.20) >Bclau.0 136227 99 + 4303871/160-280 ---------AUAGUGAGCCACA---ACUUCGGUUGUGCAAUGA-GCAAGUCAAACU----ACUUUUUAAC----GGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAA ---------.....((((((((---((....)))))((.....-))..((((((((----.((((.....----))))))))))))...)))))(((....((((.....)))).))).. ( -34.90) >Bsubt.0 9591 116 + 4214630/160-280 GGCCUGCACGACGCAGGUCACAC-AGGUGUCGCCGCAGGAUGCGGUGAACUUAACCUGUGAUCCAUUUAUC---GGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAA .....(((((.(((.((...(((-((((.(((((((.....))))))).....)))))))..))......(---((...(((((.(((((...)))))))))).)))))).))))).... ( -52.20) >Blich.0 9688 119 + 4222334/160-280 GGCCUGCACGAUGCAGGUCAUGC-AGAUGUUGUCACAGGAUGUGACGAACUUAAUCUGCGCUCCAUUAUUUCAUGGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAA (((((((.....)))))))..((-((((.(((((((.....))))))).....)))))).((((((......)))))).(((((.(((((...))))))))))...(((.....)))... ( -50.90) >consensus GGCCUGCACAAAGCAGGUCACAC_AAAUGUCGUCGCAGGAUGC_ACGAACUUAACC____AUCCAUUAAU____GGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAA ............(((.(((...........((((((.....)))))).............................((.(((((.(((((...)))))))))).))...))).))).... (-21.27 = -20.90 + -0.37)

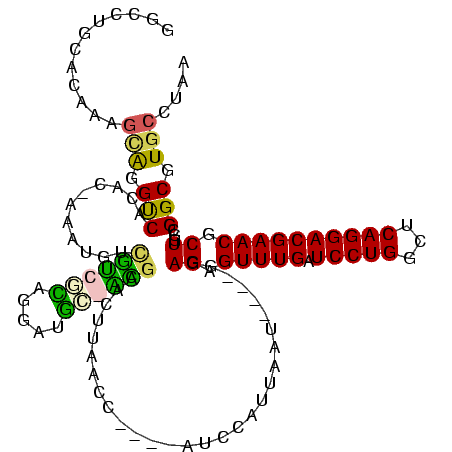
| Location | 117,184 – 117,294 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.36 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -30.16 |
| Energy contribution | -29.28 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 110 + 4202352/200-320 AAC-AUGAGCUUUAUC----AAACUUUUAU----GGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGGACCAAA-GGGAGCUUGCUCCUA ...-.((((((..(((----((((((((..----.)))))))))))...))))))((.....(((((((.(((((.......)))))..))).))))..))...-(((((....))))). ( -40.20) >Bclau.0 136227 110 + 4303871/200-320 UGA-GCAAGUCAAACU----ACUUUUUAAC----GGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGGACAGAA-GGGAGCUUGCUCCCG (((-((..((((((((----.((((.....----))))))))))))....)))))....(..(((((((.(((((.......)))))..))).))))..)....-(((((....))))). ( -41.00) >Banth.0 266645 108 + 5227293/200-320 U---GCUAGACAAACU----AA-CUUUAUU----GGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAAUGGAUUAAGAGCUUGCUCUUA .---......((((((----..-((.....----))..))))))((((((...))))))...(((((((.(((((.......)))))..))).)))).......((((((....)))))) ( -30.80) >Bsubt.0 9591 116 + 4214630/200-320 UGCGGUGAACUUAACCUGUGAUCCAUUUAUC---GGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGGACAGAU-GGGAGCUUGCUCCCU ...(((.......)))(.((.(((.......---.....(((((.(((((...))))))))))((((((.(((((.......)))))..))).)))))))).).-(((((....))))). ( -37.70) >Blich.0 9688 119 + 4222334/200-320 UGUGACGAACUUAAUCUGCGCUCCAUUAUUUCAUGGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGGACCGAC-GGGAGCUUGCUCCCU ..............(((((.((((((......)))))).(((((.(((((...))))))))))..((((.(((((.......)))))..)))).))))).....-(((((....))))). ( -40.80) >consensus UGC_GCGAGCUUAACC____AACCUUUAAU____GGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGGACAGAA_GGGAGCUUGCUCCCA .......................................(((((.(((((...))))))))))((((((.(((((.......)))))..))).))).........(((((....))))). (-30.16 = -29.28 + -0.88)

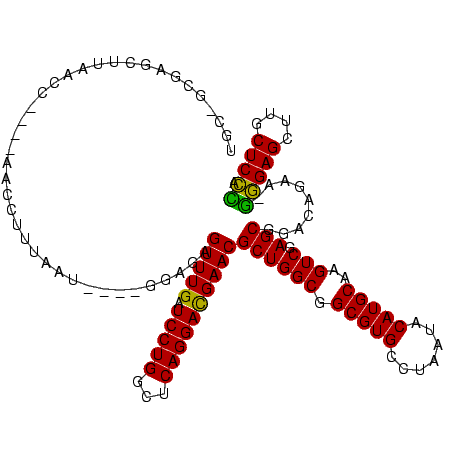
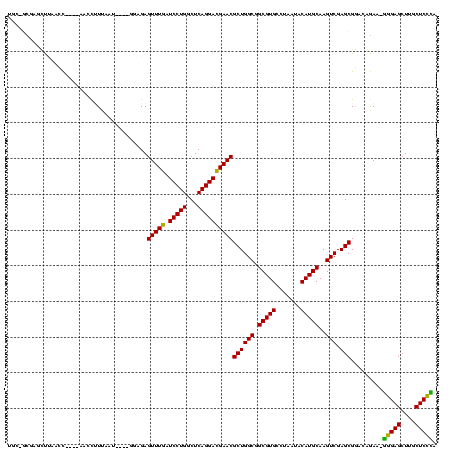
| Location | 117,184 – 117,302 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.14 |
| Mean single sequence MFE | -44.30 |
| Consensus MFE | -44.20 |
| Energy contribution | -42.72 |
| Covariance contribution | -1.48 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.95 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 118 + 4202352/280-400 UACAUGCAAGUCGAGCGGACCAAA-GGGAGCUUGCUCCUA-GAGGUUAGCGGCGAACGGGUGAGUAACACGUGGGCAACCUGCCUGUAAGACUGGGAUAACAUCGAGAAAUCGGUGCUAA ...((.(.((((..((.((((...-(((((....))))).-..)))).))((((.(((.((.....)).)))((....)))))).....)))).).))..((((((....)))))).... ( -40.60) >Bclau.0 136227 118 + 4303871/280-400 UACAUGCAAGUCGAGCGGACAGAA-GGGAGCUUGCUCCCG-GACGUUAGCGGCGGACGGGUGAGUAACACGUGGGCAACCUGCCCCUUAGACUGGGAUAACUCCGGGAAACCGGAGCUAA ...((.(.(((((((.((.....(-((..(((..(..(((-..(((.....)))..)))(((.....))))..)))..))).)).))).)))).).))..((((((....)))))).... ( -47.00) >Banth.0 266645 120 + 5227293/280-400 UACAUGCAAGUCGAGCGAAUGGAUUAAGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAA ...((.(.((((..((.(((....((((((....))))))....))).))(((((.((.(((.....))).))......))))).....)))).).))..((((((....)))))).... ( -38.90) >Bsubt.0 9591 118 + 4214630/280-400 UACAUGCAAGUCGAGCGGACAGAU-GGGAGCUUGCUCCCU-GAUGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAA ...((.(.((((..((.((((...-(((((....))))).-..)))).)).....((((((..((.((......)).))..))))))..)))).).))..((((((....)))))).... ( -46.00) >Blich.0 9688 118 + 4222334/280-400 UACAUGCAAGUCGAGCGGACCGAC-GGGAGCUUGCUCCCU-UAGGUCAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAA ...((.(.((((..((.((((...-(((((....))))).-..)))).)).....((((((..((.((......)).))..))))))..)))).).))..((((((....)))))).... ( -49.00) >consensus UACAUGCAAGUCGAGCGGACAGAA_GGGAGCUUGCUCCCA_GAGGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAA ...((.(.((((..((.((((....(((((....)))))....)))).))((((.(((.((.....)).)))((....)))))).....)))).).))..((((((....)))))).... (-44.20 = -42.72 + -1.48)

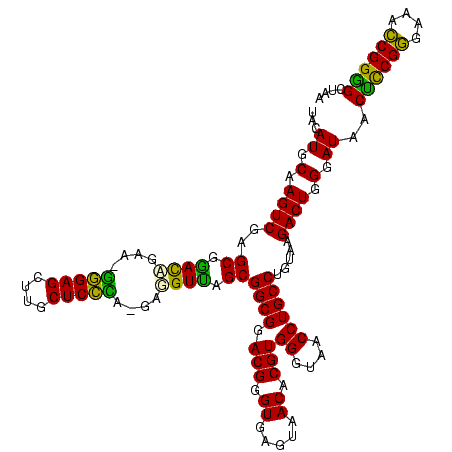
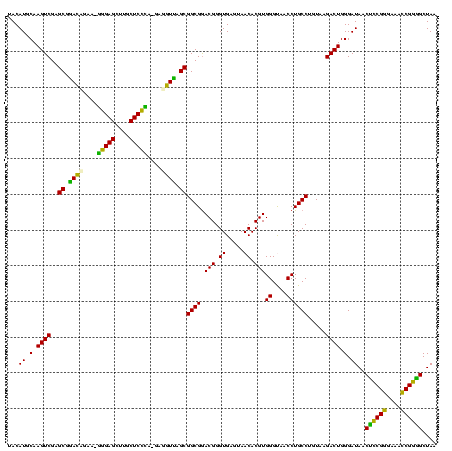
| Location | 117,184 – 117,303 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.91 |
| Mean single sequence MFE | -43.04 |
| Consensus MFE | -44.94 |
| Energy contribution | -40.34 |
| Covariance contribution | -4.60 |
| Combinations/Pair | 1.58 |
| Mean z-score | -2.82 |
| Structure conservation index | 1.04 |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.999176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 119 + 4202352/320-440 -GAGGUUAGCGGCGAACGGGUGAGUAACACGUGGGCAACCUGCCUGUAAGACUGGGAUAACAUCGAGAAAUCGGUGCUAAUACUGGAUAAUAAAAAGAACUGCAUGGUUCUUUUUUGAAA -...((((((((((.(((.((.....)).)))((....)))))).................(((((....)))))))))))..........((((((((((....))))))))))..... ( -32.60) >Bclau.0 136227 119 + 4303871/320-440 -GACGUUAGCGGCGGACGGGUGAGUAACACGUGGGCAACCUGCCCCUUAGACUGGGAUAACUCCGGGAAACCGGAGCUAAUACCGGAUAAUCCCUUUCUCCACCUGGAGAGAGGGUGAAA -..(...((.(((((.((.(((.....))).))(....)))))).))..).((((..((.((((((....)))))).))...)))).....((((((((((....))))))))))..... ( -50.80) >Banth.0 266645 120 + 5227293/320-440 UGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAA ....((((.(((....((((((.....)))((((((.....))))))....)))......((((((....))))))......)))..))))((((((((((....))))))))))..... ( -41.80) >Bsubt.0 9591 119 + 4214630/320-440 -GAUGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGCAUGGUUCAAACAUAAAA -(.((((((((((..((((((..((.((......)).))..))))))..).)))......((((((....)))))))))))).).......((((((((((....))))))))))..... ( -43.80) >Blich.0 9688 119 + 4222334/320-440 -UAGGUCAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAACCGCAUGGUUCAAUCAUAAAA -......((((.(..((((((..((.((......)).))..))))))..).((((..((.((((((....)))))).))...)))).))))((((((((((....))))))))))..... ( -46.20) >consensus _GAGGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAAUAGAUUGAACCGCAUGGUUCAAACAUGAAA ..........((((.(((.((.....)).)))((....)))))).......((((..((.((((((....)))))).))...)))).....((((((((((....))))))))))..... (-44.94 = -40.34 + -4.60)

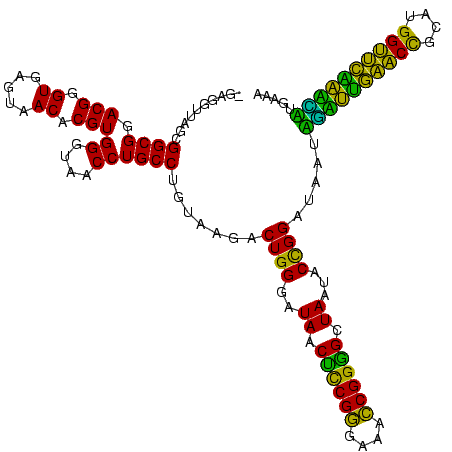
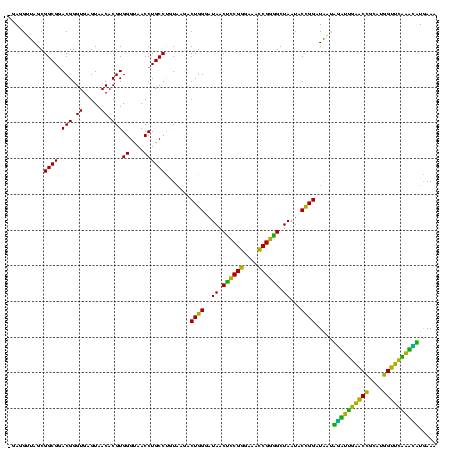
| Location | 117,184 – 117,303 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.91 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -31.78 |
| Energy contribution | -28.66 |
| Covariance contribution | -3.12 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 119 + 4202352/320-440 UUUCAAAAAAGAACCAUGCAGUUCUUUUUAUUAUCCAGUAUUAGCACCGAUUUCUCGAUGUUAUCCCAGUCUUACAGGCAGGUUGCCCACGUGUUACUCACCCGUUCGCCGCUAACCUC- .....(((((((((......)))))))))...........(((((..(((....)))...................(((.((....))..(((.....)))......))))))))....- ( -21.50) >Bclau.0 136227 119 + 4303871/320-440 UUUCACCCUCUCUCCAGGUGGAGAAAGGGAUUAUCCGGUAUUAGCUCCGGUUUCCCGGAGUUAUCCCAGUCUAAGGGGCAGGUUGCCCACGUGUUACUCACCCGUCCGCCGCUAACGUC- .....((((.(((((....))))).))))......((((..(((((((((....)))))))))............((((.....))))..(((.....)))......))))........- ( -42.20) >Banth.0 266645 120 + 5227293/320-440 UUUCAAUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUAACUUCA .....((((.(((((....))))).))))((((..((((..((((.((((....)))).))))............((((.((....))..(((.....)))..)))))))).)))).... ( -31.10) >Bsubt.0 9591 119 + 4214630/320-440 UUUUAUGUUUGAACCAUGCGGUUCAAACAACCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUAACAUC- .....((((((((((....))))))))))(((....))).(((((.((((....))))..................(((.((....))..(((.....)))......))))))))....- ( -33.60) >Blich.0 9688 119 + 4222334/320-440 UUUUAUGAUUGAACCAUGCGGUUCAAUCAAGCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUGACCUA- .....((((((((((....))))))))))(((....((...((((.((((....)))).))))..)).........(((.((....))..(((.....)))......))))))......- ( -36.70) >consensus UUUCAAGAUCGAACCAUGCGGUUCAAACAAUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUAACCUC_ .....((((((((((....))))))))))...........(((((.((((....))))..................(((.((....))..(((.....)))......))))))))..... (-31.78 = -28.66 + -3.12)

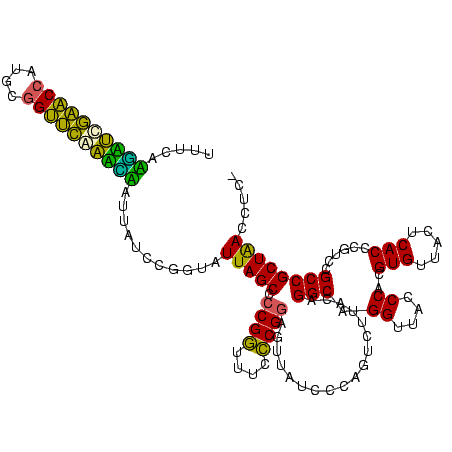
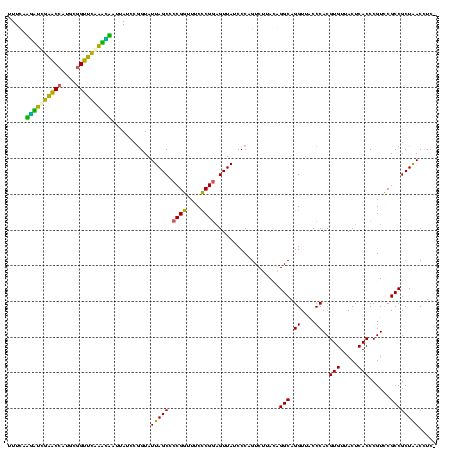
| Location | 117,184 – 117,303 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.73 |
| Mean single sequence MFE | -51.70 |
| Consensus MFE | -55.54 |
| Energy contribution | -48.38 |
| Covariance contribution | -7.16 |
| Combinations/Pair | 1.74 |
| Mean z-score | -4.29 |
| Structure conservation index | 1.07 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 119 + 4202352/360-480 UGCCUGUAAGACUGGGAUAACAUCGAGAAAUCGGUGCUAAUACUGGAUAAUAAAAAGAACUGCAUGGUUCUUUUUUGAAAGAUGGUUUC-GGCUAUCACUUACAGAUGGGCCCGCGGCGC ...(((((((.(..(..((.((((((....)))))).))...)..).....((((((((((....)))))))))).....((((((...-.)))))).)))))))....(((...))).. ( -37.00) >Bclau.0 136227 119 + 4303871/360-480 UGCCCCUUAGACUGGGAUAACUCCGGGAAACCGGAGCUAAUACCGGAUAAUCCCUUUCUCCACCUGGAGAGAGGGUGAAAGAUGGCUUC-GGCUAUCACUAGGGGAUGGGCCCGCGGCGC ...(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((...-.)))))).)))))))....(((...))).. ( -62.20) >Banth.0 266645 119 + 5227293/360-480 UGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGC ...(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((...-.)))))).)))))))...(((....))).. ( -51.30) >Bsubt.0 9591 119 + 4214630/360-480 UGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGCAUGGUUCAAACAUAAAAGGUGGCUUC-GGCUACCACUUACAGAUGGACCCGCGGCGC .(((((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((...-.)))))).))))))(.((....)))))).. ( -55.20) >Blich.0 9688 120 + 4222334/360-480 UGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAACCGCAUGGUUCAAUCAUAAAAGGUGGCUUUUAGCUACCACUUGCAGAUGGACCCGCGGCGC .(((.((......(((....((((((....))))))......(((..(((.((((((((((....)))))))))).....((((((.....))))))....)))..))).)))))))).. ( -52.80) >consensus UGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAAUAGAUUGAACCGCAUGGUUCAAACAUGAAAGGUGGCUUC_GGCUAUCACUUACAGAUGGACCCGCGGCGC .(((((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((.....)))))).)))))))..(....)))..... (-55.54 = -48.38 + -7.16)

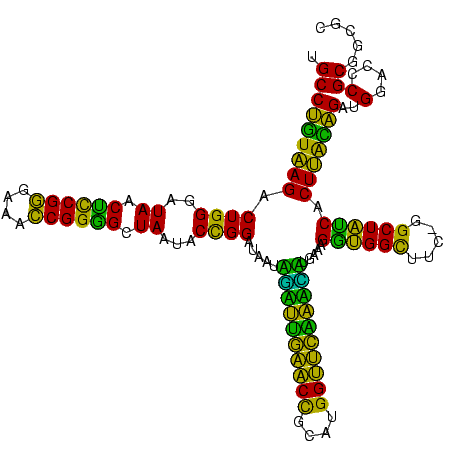
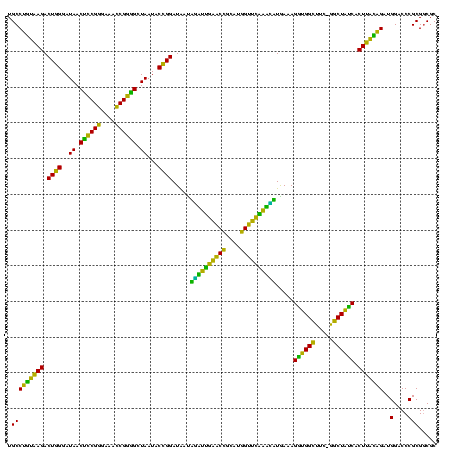
| Location | 117,184 – 117,303 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.73 |
| Mean single sequence MFE | -38.26 |
| Consensus MFE | -35.60 |
| Energy contribution | -31.60 |
| Covariance contribution | -4.00 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 119 + 4202352/360-480 GCGCCGCGGGCCCAUCUGUAAGUGAUAGCC-GAAACCAUCUUUCAAAAAAGAACCAUGCAGUUCUUUUUAUUAUCCAGUAUUAGCACCGAUUUCUCGAUGUUAUCCCAGUCUUACAGGCA (((((...)))....(((((((.(((((..-((((.....)))).(((((((((......))))))))).)))))......(((((.(((....))).))))).......))))))))). ( -27.70) >Bclau.0 136227 119 + 4303871/360-480 GCGCCGCGGGCCCAUCCCCUAGUGAUAGCC-GAAGCCAUCUUUCACCCUCUCUCCAGGUGGAGAAAGGGAUUAUCCGGUAUUAGCUCCGGUUUCCCGGAGUUAUCCCAGUCUAAGGGGCA ((.((..((((..........(.(((((..-((((.....)))).((((.(((((....))))).)))).))))))((...(((((((((....)))))))))..)).))))..)).)). ( -45.90) >Banth.0 266645 119 + 5227293/360-480 GCGACGCGGGUCCAUCCAUAAGUGACAGCC-GAAGCCGCCUUUCAAUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCA (((((....)))...((((((((((((...-..((((((..(((......)))....)))))).....)))))...((...((((.((((....)))).))))..))...))))))))). ( -35.70) >Bsubt.0 9591 119 + 4214630/360-480 GCGCCGCGGGUCCAUCUGUAAGUGGUAGCC-GAAGCCACCUUUUAUGUUUGAACCAUGCGGUUCAAACAACCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCA ..(((..((((..........(((((....-...)))))......((((((((((....))))))))))...))))((...((((.((((....)))).))))..)).........))). ( -40.20) >Blich.0 9688 120 + 4222334/360-480 GCGCCGCGGGUCCAUCUGCAAGUGGUAGCUAAAAGCCACCUUUUAUGAUUGAACCAUGCGGUUCAAUCAAGCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCA ..((((..((.(....(((..(((((........)))))......((((((((((....)))))))))).)))...((...((((.((((....)))).))))..)).).))..).))). ( -41.80) >consensus GCGCCGCGGGUCCAUCUGUAAGUGAUAGCC_GAAGCCACCUUUCAAGAUCGAACCAUGCGGUUCAAACAAUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCA (((((...)))....(((((((.(((.((.....)).))).....((((((((((....)))))))))).......((...((((.((((....)))).))))..))...))))))))). (-35.60 = -31.60 + -4.00)

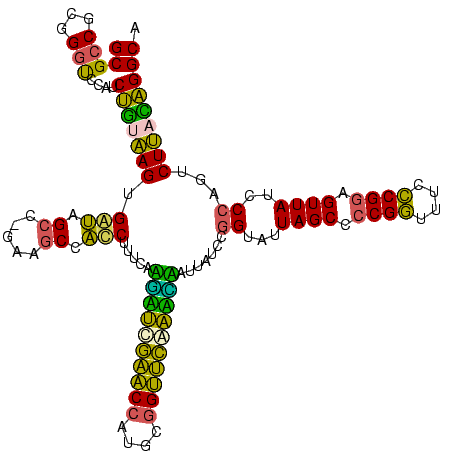
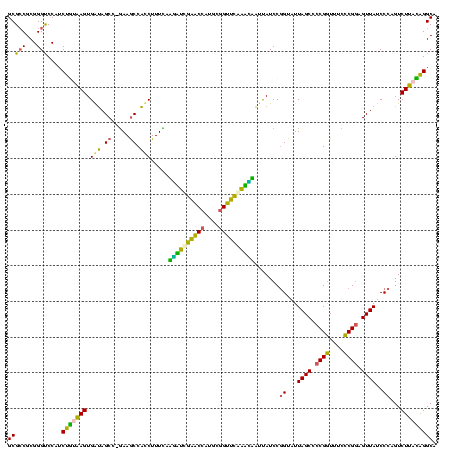
| Location | 117,184 – 117,303 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.57 |
| Mean single sequence MFE | -45.70 |
| Consensus MFE | -43.30 |
| Energy contribution | -37.98 |
| Covariance contribution | -5.32 |
| Combinations/Pair | 1.68 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 119 + 4202352/400-520 UACUGGAUAAUAAAAAGAACUGCAUGGUUCUUUUUUGAAAGAUGGUUUC-GGCUAUCACUUACAGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUGGGGUAACGGCUCACCAAGGCGACGA ..(((......((((((((((....)))))))))).....((((((...-.)))))).....)))(((.(((...))).)))..(((..((((((((.......)))))))))))..... ( -36.40) >Bclau.0 136227 119 + 4303871/400-520 UACCGGAUAAUCCCUUUCUCCACCUGGAGAGAGGGUGAAAGAUGGCUUC-GGCUAUCACUAGGGGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCAACGA (((((......((((((((((....)))))))))).....((((((...-.))))))(((((..((((.(((...))).))))..)))))))))).(((((....)))))..(....).. ( -52.30) >Banth.0 266645 119 + 5227293/400-520 UACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGA ..(((......((((((((((....)))))))))).....((((((...-.)))))).....)))((((((....))).)))..(((..((((((((.......)))))))))))..... ( -40.90) >Bsubt.0 9591 119 + 4214630/400-520 UACCGGAUGGUUGUUUGAACCGCAUGGUUCAAACAUAAAAGGUGGCUUC-GGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGA ..(((..((..((((((((((....)))))))))).....((((((...-.)))))).....))..)))...((((((......)))..((((((((.......)))))))).))).... ( -49.40) >Blich.0 9688 120 + 4222334/400-520 UACCGGAUGCUUGAUUGAACCGCAUGGUUCAAUCAUAAAAGGUGGCUUUUAGCUACCACUUGCAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGA ..(((..(((.((((((((((....)))))))))).....((((((.....))))))....)))..)))...((((((......)))..((((((((.......)))))))).))).... ( -49.50) >consensus UACCGGAUAAUAGAUUGAACCGCAUGGUUCAAACAUGAAAGGUGGCUUC_GGCUAUCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGA ..(((..((..((((((((((....)))))))))).....((((((.....)))))).....))..)))....(((((......)))..((((((((.......)))))))).))..... (-43.30 = -37.98 + -5.32)

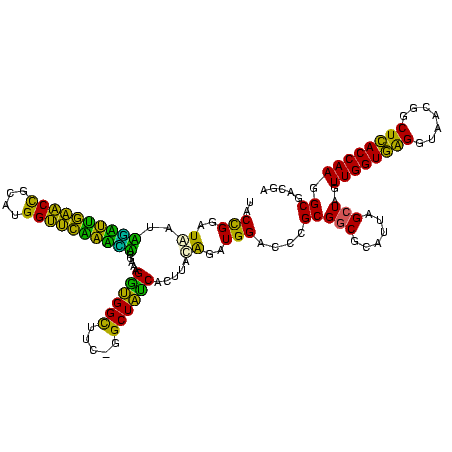
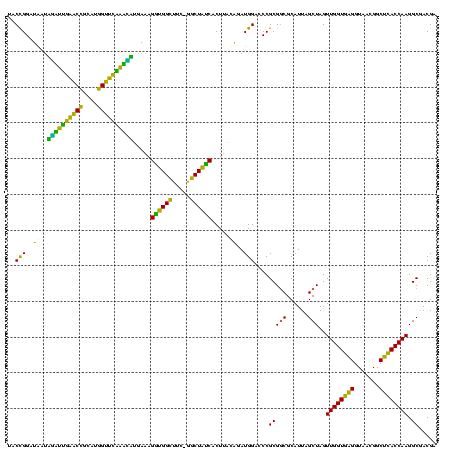
| Location | 117,184 – 117,303 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -51.54 |
| Consensus MFE | -45.86 |
| Energy contribution | -47.12 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 119 + 4202352/440-560 GAUGGUUUC-GGCUAUCACUUACAGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUGGGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((...-.)))))).....(((.((((((.((.(((((((.(((..((((((((.......)))))))))))...))))))).))..((((....))))....)))).))))).... ( -49.50) >Bclau.0 136227 119 + 4303871/440-560 GAUGGCUUC-GGCUAUCACUAGGGGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((...-.)))))).((((.(....((((.((.(((((((.(((..((((((((.......)))))))))))...))))))).))..((((....))))....)))).).))))... ( -51.60) >Banth.0 266645 119 + 5227293/440-560 GGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((...-.)))))).............(((((((((.....(((..((((((((.......)))))))))))..))))))((.((((((((....)))...))))).))...))).. ( -52.70) >Bsubt.0 9591 119 + 4214630/440-560 GGUGGCUUC-GGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((...-.)))))).............(((...(((((((.(((..((((((((.......)))))))))))...))))))).((((((((....)))...)))))......))).. ( -53.50) >Blich.0 9688 120 + 4222334/440-560 GGUGGCUUUUAGCUACCACUUGCAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((.....)))))).............(((...(((((((.(((..((((((((.......)))))))))))...))))))).((((((((....)))...)))))......))).. ( -50.40) >consensus GGUGGCUUC_GGCUAUCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((.....)))))).............(((...(((((((.(((..((((((((.......)))))))))))...))))))).((((((((....)))...)))))......))).. (-45.86 = -47.12 + 1.26)

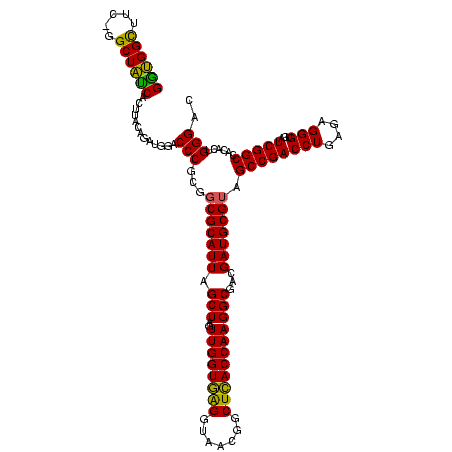
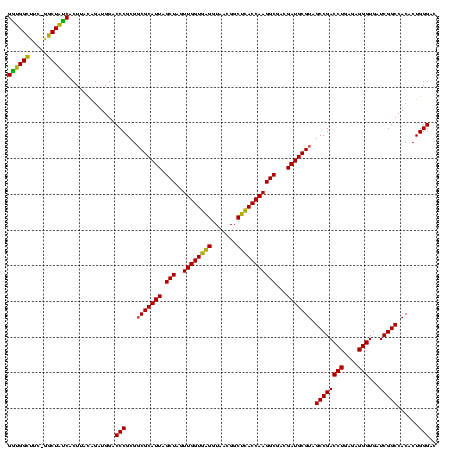
| Location | 117,184 – 117,303 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -43.10 |
| Consensus MFE | -39.36 |
| Energy contribution | -39.20 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 119 + 4202352/440-560 GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCCCACCAACUAGCUAAUGCGCCGCGGGCCCAUCUGUAAGUGAUAGCC-GAAACCAUC ......((((((((((((.........))))))))))))..(((.((.((((((...........))))))...((....)))).)))(((..(((.......))).)))-......... ( -38.50) >Bclau.0 136227 119 + 4303871/440-560 GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUGCGCCGCGGGCCCAUCCCCUAGUGAUAGCC-GAAGCCAUC ......((((((((((((.........))))))))))))..(((.((.((((((((.......))))))))...((....)))).)))(((...((..(((....)))..-)).)))... ( -41.60) >Banth.0 266645 119 + 5227293/440-560 GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCC-GAAGCCGCC (.(((.((((((((((((.........)))))))))))).((((.((.((((((((.......))))))))...))....))))...))).).........(((...((.-...))))). ( -41.50) >Bsubt.0 9591 119 + 4214630/440-560 GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGGUAGCC-GAAGCCACC (.(((.((((((((((((.........))))))))))))...((.((.((((((((.......))))))))...((....)))).))))).).........(((((....-...))))). ( -46.80) >Blich.0 9688 120 + 4222334/440-560 GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGCAAGUGGUAGCUAAAAGCCACC (.(((.((((((((((((.........))))))))))))...((.((.((((((((.......))))))))...((....)))).))))).).........(((((........))))). ( -47.10) >consensus GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGAUAGCC_GAAGCCACC (.(((.((((((((((((.........))))))))))))...((.((.((((((((.......))))))))...((....)))).))))).)...........(((.((.....)).))) (-39.36 = -39.20 + -0.16)

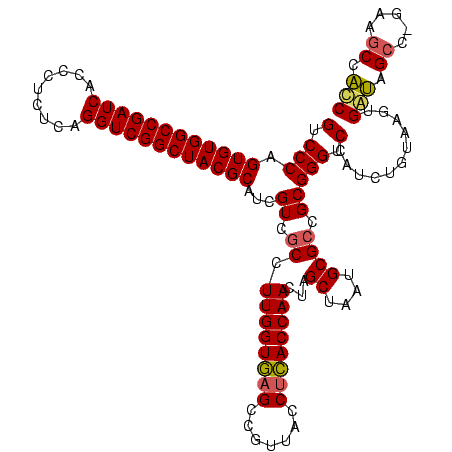
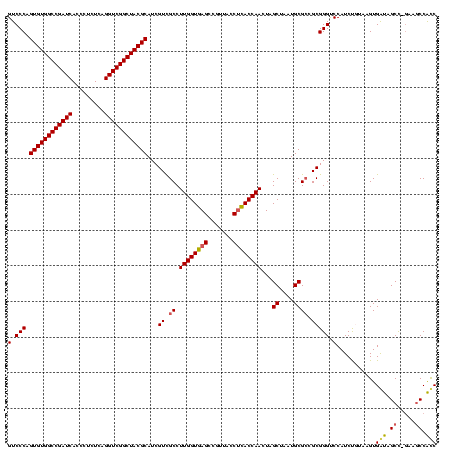
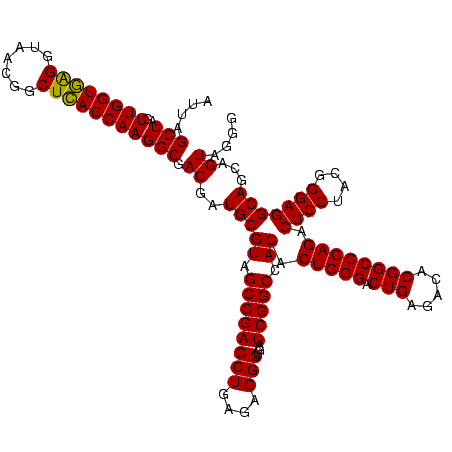
| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -51.02 |
| Consensus MFE | -46.50 |
| Energy contribution | -51.02 |
| Covariance contribution | 4.52 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/480-600 AUUAGCUAGUUGGUGGGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... ( -50.80) >Bclau.0 136227 120 + 4303871/480-600 AUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... ( -49.30) >Banth.0 266645 120 + 5227293/480-600 AUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... ( -51.60) >Bsubt.0 9591 120 + 4214630/480-600 AUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... ( -51.70) >Blich.0 9688 120 + 4222334/480-600 AUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... ( -51.70) >consensus AUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... (-46.50 = -51.02 + 4.52)


| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -41.94 |
| Energy contribution | -41.82 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.57 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/480-600 CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCCCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))......((.((((((...........))))))...)).... ( -38.90) >Bclau.0 136227 120 + 4303871/480-600 CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))...((((..((((((((.......)))))))).)))).... ( -41.10) >Banth.0 266645 120 + 5227293/480-600 CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))...((((..((((((((.......)))))))).)))).... ( -43.40) >Bsubt.0 9591 120 + 4214630/480-600 CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))......((.((((((((.......))))))))...)).... ( -43.30) >Blich.0 9688 120 + 4222334/480-600 CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))......((.((((((((.......))))))))...)).... ( -43.30) >consensus CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))......((.((((((((.......))))))))...)).... (-41.94 = -41.82 + -0.12)

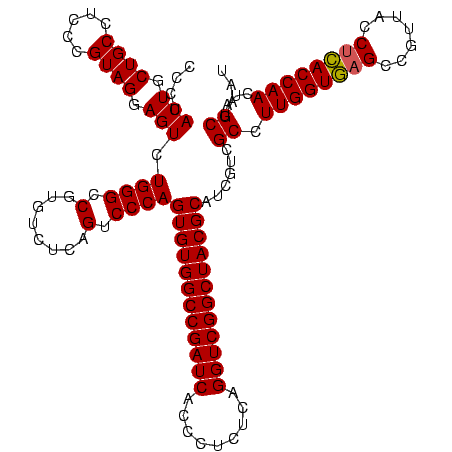
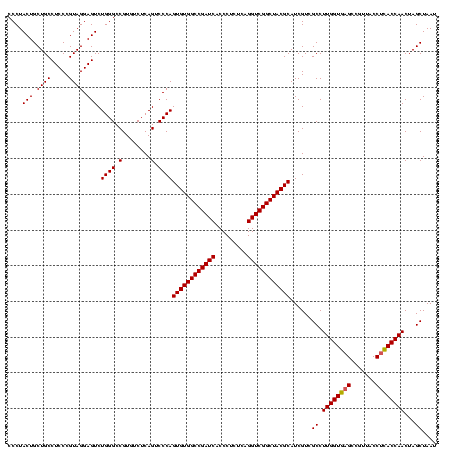
| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -49.80 |
| Consensus MFE | -44.80 |
| Energy contribution | -49.80 |
| Covariance contribution | 5.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/520-640 UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). ( -49.80) >Bclau.0 136227 120 + 4303871/520-640 UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). ( -49.80) >Banth.0 266645 120 + 5227293/520-640 UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). ( -49.80) >Bsubt.0 9591 120 + 4214630/520-640 UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). ( -49.80) >Blich.0 9688 120 + 4222334/520-640 UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). ( -49.80) >consensus UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). (-44.80 = -49.80 + 5.00)

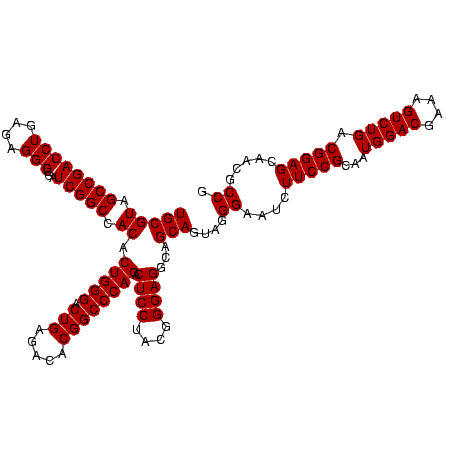
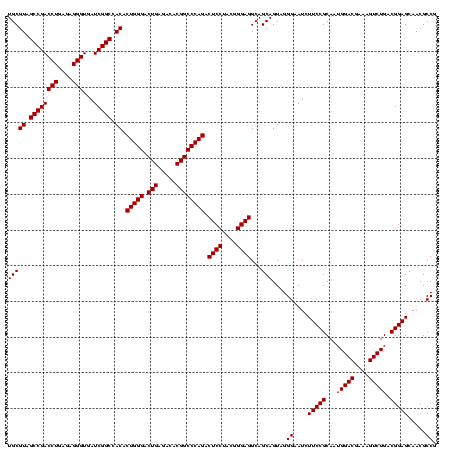
| Location | 117,184 – 117,303 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -49.52 |
| Consensus MFE | -43.90 |
| Energy contribution | -45.26 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.24 |
| Mean z-score | -4.21 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.59 |
| SVM RNA-class probability | 0.999925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 119 + 4202352/600-720 AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAA-AGGGCGGCACCUUG ..((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....)))))).((((((((((((...-.)))))))).)))) ( -49.50) >Bclau.0 136227 120 + 4303871/600-720 AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAGGAAGGCCUUCGGGUCGUAAAGCUCUGUUGUGAGGGAAGAAGCGGUACCGUUCGAAUAGGGCGGUACCUUG ..((((((.((..((((....))))((((((((.....(((....))).....((((....)))).....)))))))).))..))))))...((((((((((.....))))))))))... ( -58.80) >Banth.0 266645 120 + 5227293/600-720 AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUG ..((((((.....((((....))))(((((((..(((((.((.((((.......)))).)))).)))....))))))).....)))))).((((((((((((.....)))))))).)))) ( -45.90) >Bsubt.0 9591 120 + 4214630/600-720 AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAAUAGGGCGGUACCUUG ..((((((.....((((....))))((((((((..((.(((....))).))..((((....)))).....)))))))).....)))))).((((((((((((.....)))))))).)))) ( -48.90) >Blich.0 9688 120 + 4222334/600-720 AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUG ..((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....)))))).((((((((((((.....)))))))).)))) ( -44.50) >consensus AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAAUAGGGCGGUACCUUG ..((((((.....((((....))))(((((((...(..(((....)))..)..((((....))))......))))))).....))))))....(((((((((.....))))))))).... (-43.90 = -45.26 + 1.36)

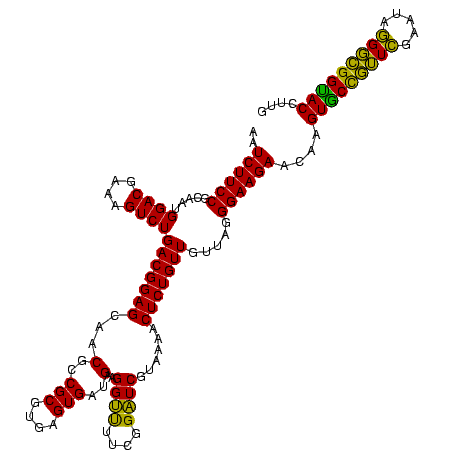
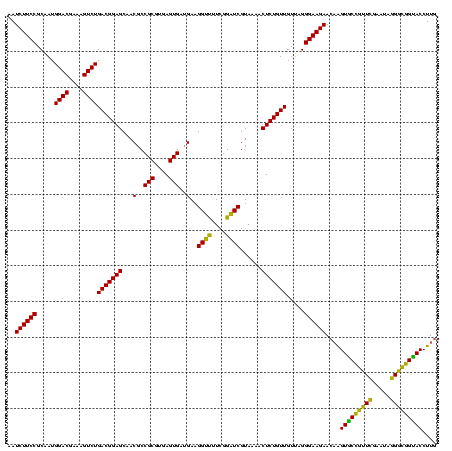
| Location | 117,184 – 117,303 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -41.39 |
| Consensus MFE | -38.58 |
| Energy contribution | -37.06 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.996490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 119 + 4202352/640-760 CGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAA-AGGGCGGCACCUUGACGGUACCUAACGAGAAAGCCACGGCUAACUACGUGCCAG (....).......((((((((((.........))).((((((((......((((((((((((...-.)))))))).))))......)))))))))))))))..(((.........))).. ( -43.40) >Bclau.0 136227 120 + 4303871/640-760 CGUGAGUGAGGAAGGCCUUCGGGUCGUAAAGCUCUGUUGUGAGGGAAGAAGCGGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUCACCAGAAAGCCACGGCUAACUACGUGCCAG .(((.((......((((....)))).......((((..((((((........((((((((((.....)))))))))).........))))))))))..)))))(((.........))).. ( -46.53) >Banth.0 266645 120 + 5227293/640-760 CGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAG (....).......(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..)))))))..(((.........))).. ( -40.90) >Bsubt.0 9591 120 + 4214630/640-760 CGUGAGUGAUGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAG .(((.((......((((...(((((((....(((((....))))).....((((((((((((.....)))))))).))))))))).)).)))).....)))))(((.........))).. ( -37.90) >Blich.0 9688 120 + 4222334/640-760 CGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAG .(((.((......((((...(((((((....(((((....))))).....((((((((((((.....)))))))).))))))))).)).)))).....)))))(((.........))).. ( -38.20) >consensus CGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAG .(((.((......((((....)))).......((((..((((((......((((((((((((.....)))))))).))))......))))))))))..)))))(((.........))).. (-38.58 = -37.06 + -1.52)

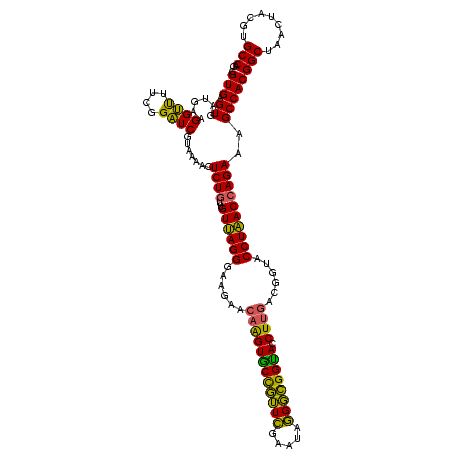
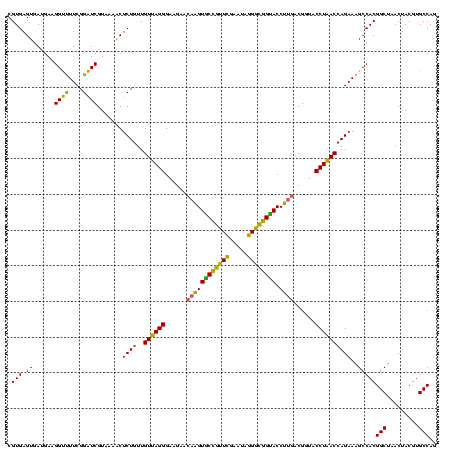
| Location | 117,184 – 117,303 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -42.83 |
| Consensus MFE | -42.06 |
| Energy contribution | -40.58 |
| Covariance contribution | -1.48 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.19 |
| SVM RNA-class probability | 0.998697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 119 + 4202352/680-800 UAGGGAAGAACAAGUGCCGUUCGAA-AGGGCGGCACCUUGACGGUACCUAACGAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAU ((((......((((((((((((...-.)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)))).... ( -45.60) >Bclau.0 136227 120 + 4303871/680-800 GAGGGAAGAAGCGGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUCACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAU ((((........((((((((((.....)))))))))).........)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... ( -45.03) >Banth.0 266645 120 + 5227293/680-800 UAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUAUCCGGAAU ((((......((((((((((((.....)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... ( -40.70) >Bsubt.0 9591 120 + 4214630/680-800 UAGGGAAGAACAAGUGCCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAU ((((......((((((((((((.....)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... ( -41.40) >Blich.0 9688 120 + 4222334/680-800 UAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAU ((((......((((((((((((.....)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... ( -41.40) >consensus UAGGGAAGAACAAGUGCCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAU ((((......((((((((((((.....)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... (-42.06 = -40.58 + -1.48)

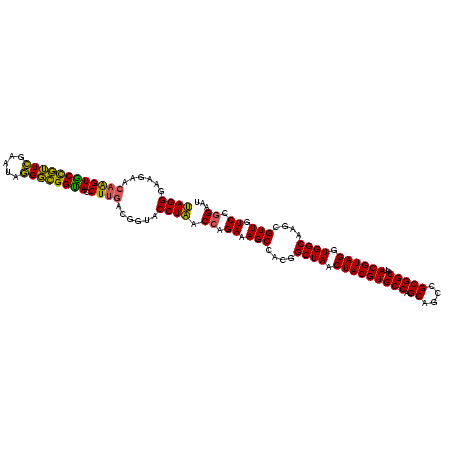

| Location | 117,184 – 117,303 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -39.31 |
| Consensus MFE | -36.34 |
| Energy contribution | -36.46 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 119 + 4202352/680-800 AUUCCGGACAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUCGUUAGGUACCGUCAAGGUGCCGCCCU-UUCGAACGGCACUUGUUCUUCCCUA .....((((((..(((((.(((((((.....((((((((((((.....))).)))))))))......)).)))))...).))))((((((..(.-...)..))))))))))))....... ( -40.50) >Bclau.0 136227 120 + 4303871/680-800 AUUCCGGACAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUGAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGUACCGCUUCUUCCCUC .....(((....((....((((.........((((((((((((.....))).)))))))))......)))).(((((((((.(((......)))....).))))))))))....)))... ( -39.66) >Banth.0 266645 120 + 5227293/680-800 AUUCCGGAUAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCAGCUUAUUCAACUAGCACUUGUUCUUCCCUA .....(((.((((..(((.............((((((((((((.....))).)))))))))......((((.((((((.....))))))))))..........)))..))))..)))... ( -37.20) >Bsubt.0 9591 120 + 4214630/680-800 AUUCCGGACAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGCACUUGUUCUUCCCUA .....(((.((((..((((............((((((((((((.....))).)))))))))......(((..((((((.....)))))))))..........))))..))))..)))... ( -40.60) >Blich.0 9688 120 + 4222334/680-800 AUUCCGGACAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGUACUUGUUCUUCCCUA .....(((.((((..((((............((((((((((((.....))).)))))))))......(((..((((((.....)))))))))..........))))..))))..)))... ( -38.60) >consensus AUUCCGGACAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGCACUUGUUCUUCCCUA .....(((.((((..((((............((((((((((((.....))).)))))))))...........((((((.....)))))).............))))..))))..)))... (-36.34 = -36.46 + 0.12)


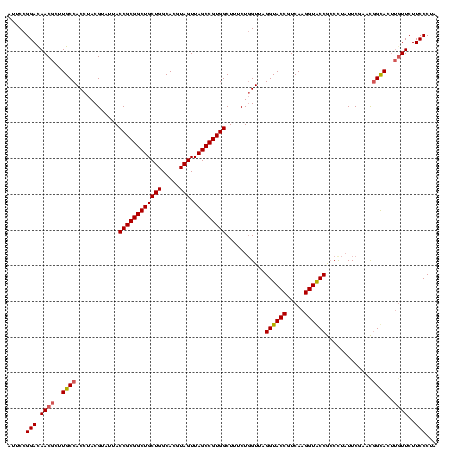
| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -39.92 |
| Consensus MFE | -38.82 |
| Energy contribution | -36.74 |
| Covariance contribution | -2.08 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.743022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/800-920 UAUUGGGCGUAAAGCGCGCGCAGGCGGUCUCUUAAGUCUGAUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGAGACUUGAGUACAGAAGAGGAGAGUGGAAUUCCA ..(((.((((.....)))).))).((.((((((...((((..((((....((((((.....))))))....)))).....(((((....))..))).))))..)))))).))........ ( -40.80) >Bclau.0 136227 120 + 4303871/800-920 UAUUGGGCGUAAAGCGCGCGCAGGCGGCUUCUUAAGUCUGAUGUGAAAUCUCGGGGCUCAACCCCGAGCGGCCAUUGGAAACUGGGGAGCUUGAGUGCAGAAGAGGAGAGUGGAAUUCCA ....(.(((.....))).)(((..((((((((((..((..(((.(....(((((((.....)))))))...))))..))...)))))))).))..)))......((((.......)))). ( -43.70) >Banth.0 266645 120 + 5227293/800-920 UAUUGGGCGUAAAGCGCGCGCAGGUGGUUUCUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCA ......(((.....)))((((....(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))...))))......((((.......)))). ( -38.10) >Bsubt.0 9591 120 + 4214630/800-920 UAUUGGGCGUAAAGGGCUCGCAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCA ....((((.......))))(((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))......((((.......)))). ( -37.80) >Blich.0 9688 120 + 4222334/800-920 UAUUGGGCGUAAAGCGCGCGCAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCA ....(.(((.....))).)(((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))......((((.......)))). ( -39.20) >consensus UAUUGGGCGUAAAGCGCGCGCAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCA ....(.(((.....))).)(((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))......((((.......)))). (-38.82 = -36.74 + -2.08)

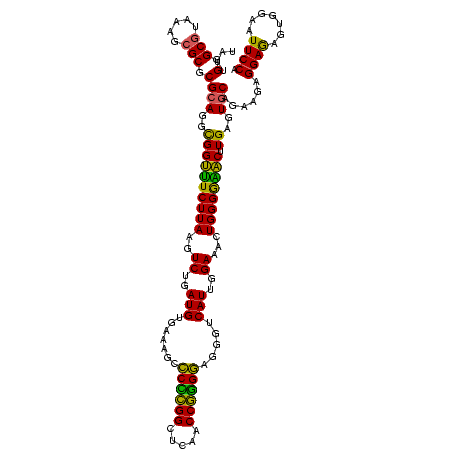
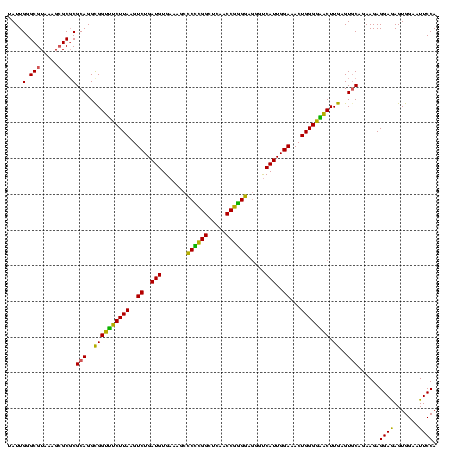
| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -34.98 |
| Energy contribution | -32.94 |
| Covariance contribution | -2.04 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/840-960 AUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGAGACUUGAGUACAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCA .((((.....((((((.....)))))).(((((...(....).....)))))...))))........(.(((...((((((((.(.(((......)))....).))))))))...)))). ( -36.70) >Bclau.0 136227 120 + 4303871/840-960 AUGUGAAAUCUCGGGGCUCAACCCCGAGCGGCCAUUGGAAACUGGGGAGCUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCA ........((((((((.....))))((((..((...(....)..))..)))).............))))(((...((((((((.(.(((......)))....).))))))))...))).. ( -39.00) >Banth.0 266645 120 + 5227293/840-960 AUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCA ........((((((((.....)))))).(((((...(....).....)))))....))...........(((...((((((((.(.(((......)))....).))))))))...))).. ( -32.20) >Bsubt.0 9591 120 + 4214630/840-960 AUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCA ........((((((((.....))))))((..((...(....)...))..)).....)).........(.(((...((((((((.(.(((......)))....).))))))))...)))). ( -35.80) >Blich.0 9688 120 + 4222334/840-960 AUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCA ........((((((((.....))))))((..((...(....)...))..)).....)).........(.(((...((((((((.(.(((......)))....).))))))))...)))). ( -35.80) >consensus AUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCA .((((.....((((((.....))))))((..((...(....)...))..))....))))..........(((...((((((((.(.(((......)))....).))))))))...))).. (-34.98 = -32.94 + -2.04)

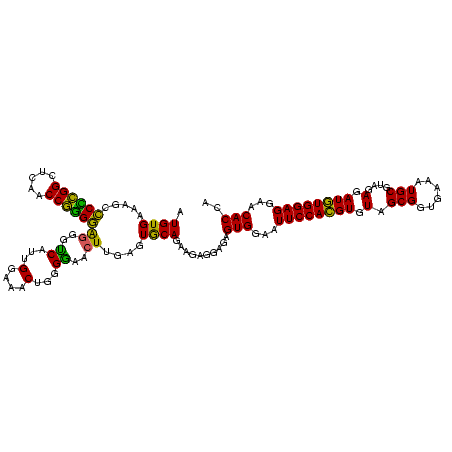
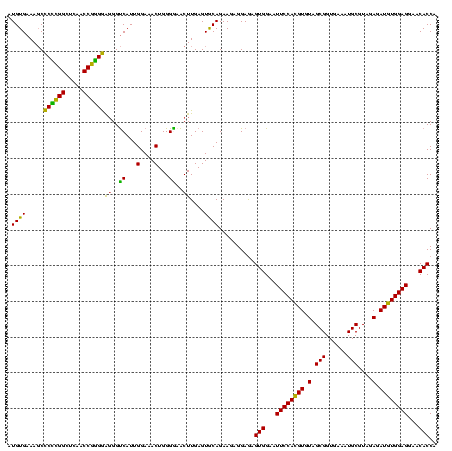
| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -24.72 |
| Energy contribution | -23.24 |
| Covariance contribution | -1.48 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/840-960 UGGUGUUCCUCCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGUACUCAAGUCUCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAU .(((((......)))))..(((((........)).....((((....))))...........))).....................(((...(((((((.....)))))))...)))... ( -25.30) >Bclau.0 136227 120 + 4303871/840-960 UGGUGUUCCUCCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGCUCCCCAGUUUCCAAUGGCCGCUCGGGGUUGAGCCCCGAGAUUUCACAU .((((.......................))))((((...((((....))))...........((......))..............))))..(((((((.....)))))))......... ( -27.70) >Banth.0 266645 120 + 5227293/840-960 UGGUGUUCCUCCAUAUCUCUACGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAU .((((.......................))))((.......((((.......))))......))......................(((...(((((((.....)))))))...)))... ( -21.52) >Bsubt.0 9591 120 + 4214630/840-960 UGGUGUUCCUCCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUUCCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAU .(((((................((........)).....((((....))))...........)))))...................(((...(((((((.....)))))))...)))... ( -25.40) >Blich.0 9688 120 + 4222334/840-960 UGGUGUUCCUCCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUUCCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAU .(((((................((........)).....((((....))))...........)))))...................(((...(((((((.....)))))))...)))... ( -25.40) >consensus UGGUGUUCCUCCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUUCCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAU .(((((................((........)).....((((....))))...........)))))...................(((...(((((((.....)))))))...)))... (-24.72 = -23.24 + -1.48)

| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -40.06 |
| Consensus MFE | -40.08 |
| Energy contribution | -39.28 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.51 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.71 |
| SVM RNA-class probability | 0.999941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/880-1000 ACUGGGAGACUUGAGUACAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGG .((.((.....(.((((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).)..)).)). ( -41.00) >Bclau.0 136227 120 + 4303871/880-1000 ACUGGGGAGCUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGG .......(((.(.((((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).).))).... ( -42.90) >Banth.0 266645 120 + 5227293/880-1000 ACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGG .(.((....)).)((((((((..(((((.(((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).......... ( -36.40) >Bsubt.0 9591 120 + 4214630/880-1000 ACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGG .((.((...(.(.((((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).).))).)). ( -40.00) >Blich.0 9688 120 + 4222334/880-1000 ACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGG .((.((...(.(.((((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).).))).)). ( -40.00) >consensus ACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGG .((.((.....(.((((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).)..)).)). (-40.08 = -39.28 + -0.80)


| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -44.64 |
| Consensus MFE | -45.52 |
| Energy contribution | -44.20 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.42 |
| Structure conservation index | 1.02 |
| SVM decision value | 5.08 |
| SVM RNA-class probability | 0.999972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/1000-1120 CGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGAUGCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCC .(((...(((((((..((...((((.........))))))..)))))))))).......(((((((.((..((((((((......))))))))..))...((....))....))))))). ( -44.30) >Bclau.0 136227 120 + 4303871/1000-1120 CGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGAUGCCCGUAGUGCCGAAGUUAACACAUUAAGCACUCC .(((...(((((((..((...((((.........))))))..)))))))))).......(((((((.((..(((.((((......)))).)))..))...((....))....))))))). ( -42.60) >Banth.0 266645 120 + 5227293/1000-1120 CGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCC .(((...(((((((..((...((((.........))))))..)))))))))).......(((((((.((..(((((((((....)))))))))..))...((....))....))))))). ( -42.40) >Bsubt.0 9591 120 + 4214630/1000-1120 AGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCC .(((...((((((...((...((((.........))))...))))))))))).......(((((((.((..(((((((((....)))))))))..))...((....))....))))))). ( -48.90) >Blich.0 9688 120 + 4222334/1000-1120 CGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGCAGCAAACGCAUUAAGCACUCC .(((...((((((...((...((((.........))))...))))))))))).......(((((((.((..(((((((((....)))))))))..))...((....))....))))))). ( -45.00) >consensus CGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGAAGUUAACGCAUUAAGCACUCC .(((...(((((((..((...((((.........))))))..)))))))))).......(((((((.((..((((((((......))))))))..))...((....))....))))))). (-45.52 = -44.20 + -1.32)

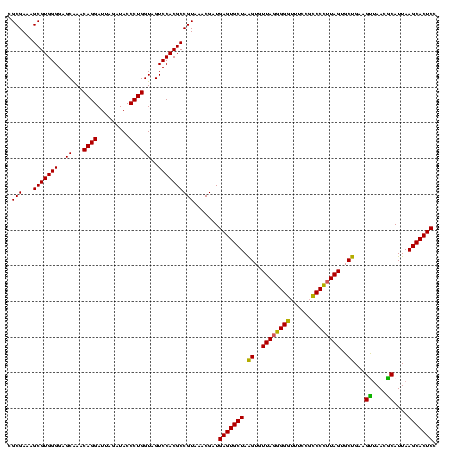
| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -38.44 |
| Consensus MFE | -32.66 |
| Energy contribution | -32.34 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/1000-1120 GGAGUGCUUAAUGUGUUAACUUCGGCACUAAGGGCAUCGAAACCCCUAACACCUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCG .(((((((....((((((.(....)......(((........))).))))))..)))))))..(((....(((((((.....((((((.....))))))........)))))))...))) ( -38.12) >Bclau.0 136227 120 + 4303871/1000-1120 GGAGUGCUUAAUGUGUUAACUUCGGCACUACGGGCAUCGAAACCCCUAACACCUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCG .(((((((....((((((..((((((.(....)))..)))).....))))))..)))))))..(((....(((((((.....((((((.....))))))........)))))))...))) ( -38.52) >Banth.0 266645 120 + 5227293/1000-1120 GGAGUGCUUAAUGCGUUAACUUCAGCACUAAAGGGCGGAAACCCUCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCG .(((((((((.(((..........))).)))((((.(....).))))........))))))..(((....(((((((.....((((((.....))))))........)))))))...))) ( -35.72) >Bsubt.0 9591 120 + 4214630/1000-1120 GGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUCGCUCCCCACGCUUUCGCU .(((((((....(.((((((....)).....((((.(....))))))))).)..))))))).........(((((((.....((((((.....))))))........)))))))...... ( -42.72) >Blich.0 9688 120 + 4222334/1000-1120 GGAGUGCUUAAUGCGUUUGCUGCAGCACUAAAGGGCGGAAACCCUCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUCGCUCCCCACGCUUUCGCG .(((((((....(.((((((....)))....((((.(....).))))))).)..)))))))..(((....(((((((.....((((((.....))))))........)))))))...))) ( -37.12) >consensus GGAGUGCUUAAUGCGUUAACUUCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCG .(((((((...((.(((......))).....((((......))))....))...)))))))...((....(((((((.....((((((.....))))))........)))))))...)). (-32.66 = -32.34 + -0.32)

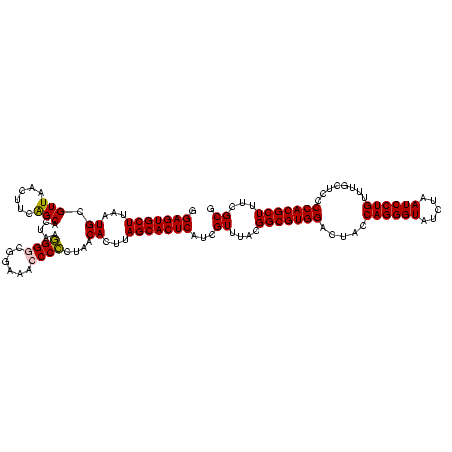
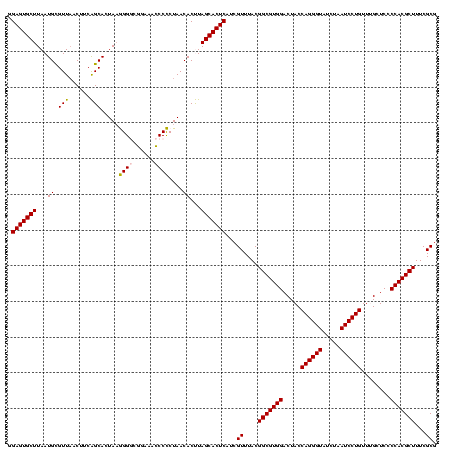
| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.17 |
| Mean single sequence MFE | -47.68 |
| Consensus MFE | -49.48 |
| Energy contribution | -47.36 |
| Covariance contribution | -2.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.43 |
| Structure conservation index | 1.04 |
| SVM decision value | 6.32 |
| SVM RNA-class probability | 0.999998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/1040-1160 AGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGAUGCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..((((((((......))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ ( -46.70) >Bclau.0 136227 120 + 4303871/1040-1160 AGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGAUGCCCGUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..(((.((((......)))).)))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ ( -47.00) >Banth.0 266645 120 + 5227293/1040-1160 AGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ ( -46.80) >Bsubt.0 9591 120 + 4214630/1040-1160 AGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ ( -50.90) >Blich.0 9688 120 + 4222334/1040-1160 AGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGCAGCAAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ ( -47.00) >consensus AGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..((((((((......))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ (-49.48 = -47.36 + -2.12)

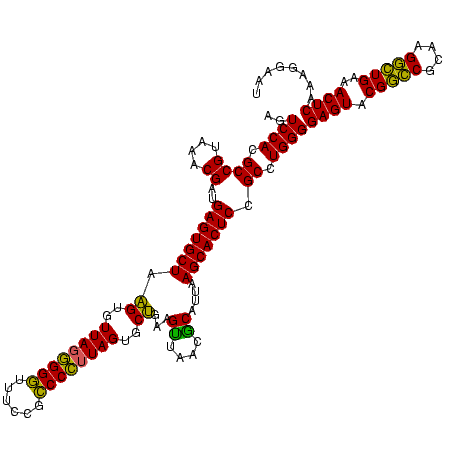
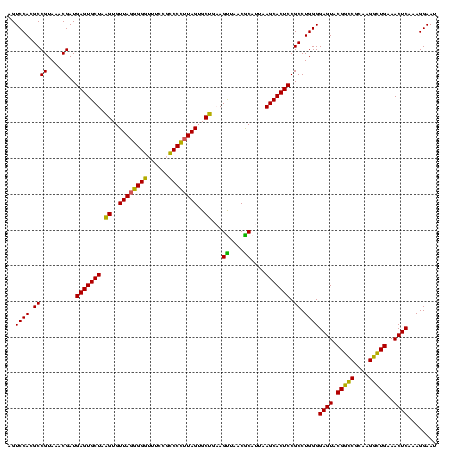
| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.83 |
| Mean single sequence MFE | -42.64 |
| Consensus MFE | -38.08 |
| Energy contribution | -38.16 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/1080-1200 UCGAUGCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCAGUGGAGCAUGUGGUUUAAU .....(((((((((((..((((....)).))..)))))...(((...((((.(((((....)))))..))))..))).......))))))...((((.((......)).))))....... ( -37.40) >Bclau.0 136227 120 + 4303871/1080-1200 UCGAUGCCCGUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCAGUGGAGCAUGUGGUUUAAU ......((((((((((..((((....)).))..)))))...(((...((((.(((((....)))))..))))..))).....)))))......((((.((......)).))))....... ( -38.80) >Banth.0 266645 120 + 5227293/1080-1200 UUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAU ((((((((...((((((...((....))....))))))((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).))))............. ( -44.00) >Bsubt.0 9591 120 + 4214630/1080-1200 UUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAU ((((((((((.((((((...((....))....))))))...(((...((((.(((((....)))))..))))..))).......))))))((((....)))).))))............. ( -48.80) >Blich.0 9688 120 + 4222334/1080-1200 UUCCGCCCUUUAGUGCUGCAGCAAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAU ((((((((...((((((...((....))....))))))((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).))))............. ( -44.20) >consensus UUCCGCCCCUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAU ....(((((..((((((...((....))....))))))...(((...((((.(((((....)))))..))))..)))........)))))...((((.((......)).))))....... (-38.08 = -38.16 + 0.08)

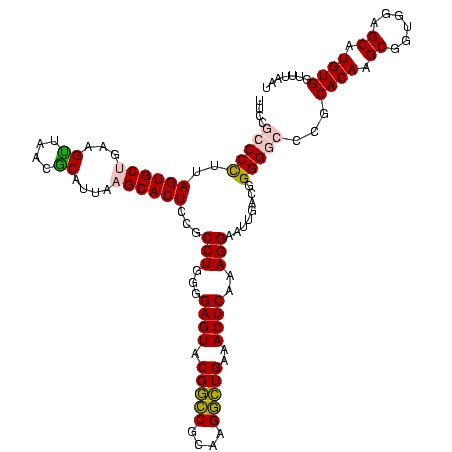
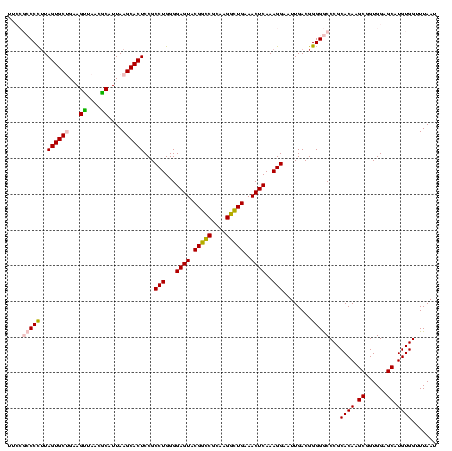
| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -39.88 |
| Consensus MFE | -40.68 |
| Energy contribution | -39.88 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.07 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/1120-1240 GCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCAGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAGGUCUUGACAUCCU ((((((.((((.(((((....)))))..)))).((((......(((....)))((((.((......)).))))......((((........))))...)))).))))))........... ( -38.70) >Bclau.0 136227 120 + 4303871/1120-1240 GCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCAGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAGGUCUUGACAUCCU ((((((.((((.(((((....)))))..)))).((((......(((....)))((((.((......)).))))......((((........))))...)))).))))))........... ( -40.70) >Banth.0 266645 120 + 5227293/1120-1240 GCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAGGUCUUGACAUCCU ((((((.((((.(((((....)))))..)))).((((......(((....)))((((.((......)).))))......((((........))))...)))).))))))........... ( -41.40) >Bsubt.0 9591 120 + 4214630/1120-1240 GCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAGGUCUUGACAUCCU ((((((.((((.(((((....)))))..)))).((((......(((....)))((((.((......)).))))......((((........))))...)))).))))))........... ( -39.30) >Blich.0 9688 120 + 4222334/1120-1240 GCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAGGUCUUGACAUCCU ((((((.((((.(((((....)))))..)))).((((......(((....)))((((.((......)).))))......((((........))))...)))).))))))........... ( -39.30) >consensus GCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAGGUCUUGACAUCCU ((((((.((((.(((((....)))))..)))).((((......(((....)))((((.((......)).))))......((((........))))...)))).))))))........... (-40.68 = -39.88 + -0.80)

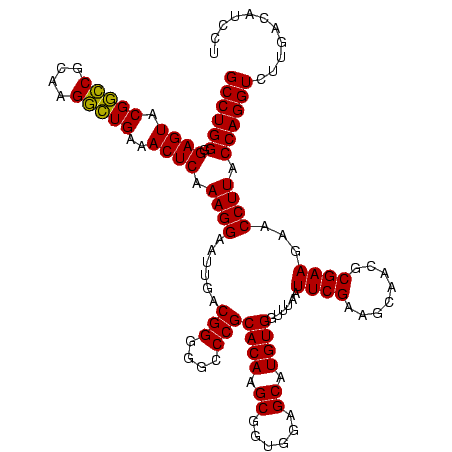
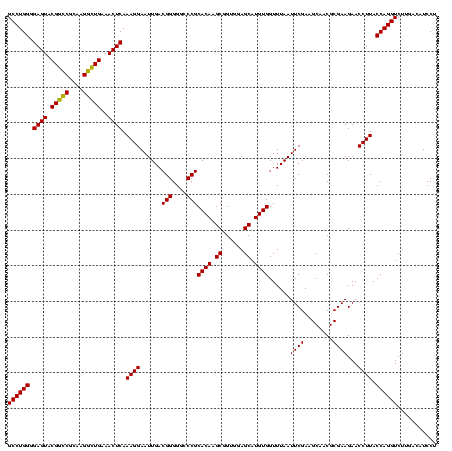
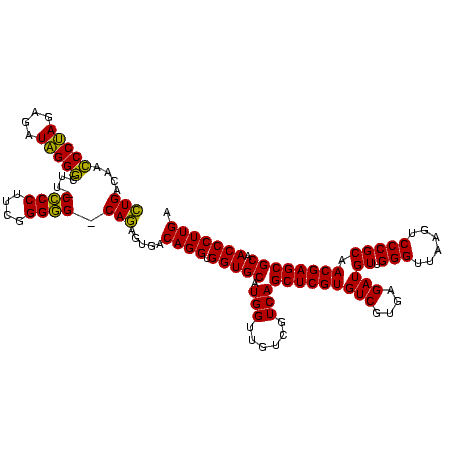
| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.78 |
| Mean single sequence MFE | -45.82 |
| Consensus MFE | -39.82 |
| Energy contribution | -44.38 |
| Covariance contribution | 4.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/1240-1360 UUGACCACCCUAGAGAUAGGGCUUUCCCCUUCGGGGGACAAAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUGA ...(((((((((....))).(((((((((....)))))..))))....)))))).((.((((((.....(((((((((....)))((.(((......))))).))))))))))))..)). ( -48.90) >Bclau.0 136227 118 + 4303871/1240-1360 UUGAUCACCCAAGAGAUUGGGCUU-CCCCUUCGGGGG-CAAAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUGA ....((((((((....))))....-((((....))))-....))))((((.(((((.(((......)))(((((((((....)))((.(((......))))).)))))))).))))))). ( -44.70) >Banth.0 266645 118 + 5227293/1240-1360 CUGACAACCCUAGAGAUAGGGCUU-CUCCUUCGGGAG-CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUGA (((....(((((....)))))...-((((....))))-))).....((((.(((((.(((......)))(((((((((....)))((.(((......))))).)))))))).))))))). ( -44.20) >Bsubt.0 9591 118 + 4214630/1240-1360 CUGACAAUCCUAGAGAUAGGACGU-CCCCUUCGGGGG-CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUGA (((....(((((....)))))...-((((....))))-))).....((((.(((((.(((......)))(((((((((....)))((.(((......))))).)))))))).))))))). ( -44.80) >Blich.0 9688 118 + 4222334/1240-1360 CUGGCAACCCUAGAGAUAGGGCUU-CCCCUUCGGGGG-CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUGA (((....(((((....)))))...-((((....))))-))).....((((.(((((.(((......)))(((((((((....)))((.(((......))))).)))))))).))))))). ( -46.50) >consensus CUGACAACCCUAGAGAUAGGGCUU_CCCCUUCGGGGG_CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUGA (((....(((((....)))))....((((....)))).))).....((((.(((((.(((......)))(((((((((....)))((.(((......))))).)))))))).))))))). (-39.82 = -44.38 + 4.56)

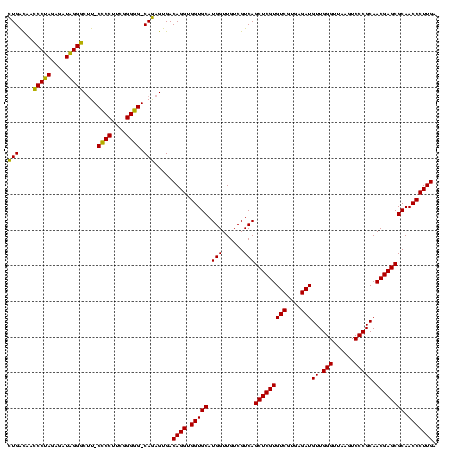
| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -39.86 |
| Consensus MFE | -37.00 |
| Energy contribution | -36.32 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/1400-1520 GCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUGACCUGGGCUACACACGUGCUACAAUGGAUGGUACAAAGGGUUGCGAAGCCGCGAGGUGAAGCCA .(((((.....)))))(((..(..((((((((.(......).)))).))))...)..))).((((.(((..((((((.......))))))......(((((....))))).))).)))). ( -41.80) >Bclau.0 136227 120 + 4303871/1400-1520 GCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUGACCUGGGCUACACACGUGCUACAAUGGAUGGUACAAAGGGCAGCGAAACCGCGAGGUGAAGCCA .(((((.....)))))(((..(..((((((((.(......).)))).))))...)..))).((((.(((..((((((.......))))))........(((....)))...))).)))). ( -40.50) >Banth.0 266645 120 + 5227293/1400-1520 GCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUGACCUGGGCUACACACGUGCUACAAUGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUA .(((((.....))))).((.((((((((((((.(......).)))).)))).....))))...))..((.(((.(......).)))))......((((.(...(((....)))).)))). ( -36.40) >Bsubt.0 9591 120 + 4214630/1400-1520 GCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUGACCUGGGCUACACACGUGCUACAAUGGACAGAACAAAGGGCAGCGAAACCGCGAGGUUAAGCCA .(((((.....)))))(((..(..((((((((.(......).)))).))))...)..))).((((.....((((((....((.......))...)))).)).((((....)))).)))). ( -39.10) >Blich.0 9688 120 + 4222334/1400-1520 GCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUGACCUGGGCUACACACGUGCUACAAUGGGCAGAACAAAGGGCAGCGAAGCCGCGAGGCUAAGCCA .(((((.....)))))(((..(..((((((((.(......).)))).))))...)..))).((((.....((((((....((.......))...)))).)).((((....)))).)))). ( -41.50) >consensus GCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUGACCUGGGCUACACACGUGCUACAAUGGACGGUACAAAGGGCAGCGAAACCGCGAGGUGAAGCCA .(((((.....)))))(((..(..((((((((.(......).)))).))))...)..))).((((.....((((((....((.......))...))).)))..(((....)))..)))). (-37.00 = -36.32 + -0.68)

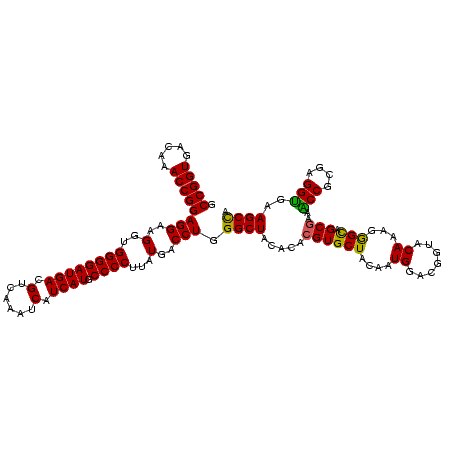
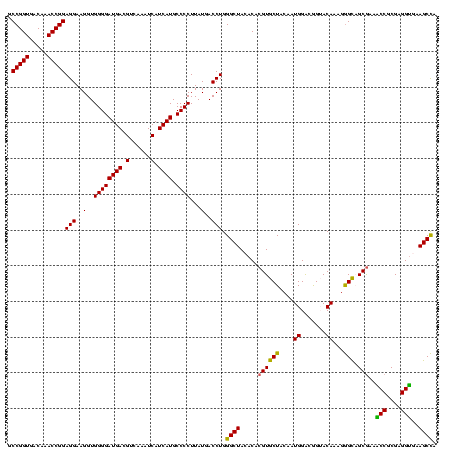
| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.00 |
| Mean single sequence MFE | -41.62 |
| Consensus MFE | -43.86 |
| Energy contribution | -39.70 |
| Covariance contribution | -4.16 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.74 |
| Structure conservation index | 1.05 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/1480-1600 UGGAUGGUACAAAGGGUUGCGAAGCCGCGAGGUGAAGCCAAUCCCAGAAAGCCAUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAAGCCGGAAUUGCUAGUAAUCGCGGAUC .(((((((.....(((((((...(((....)))...).))))))......)))))))....(((((..(((((((........))))))).....))))).((((.(....).))))... ( -42.80) >Bclau.0 136227 120 + 4303871/1480-1600 UGGAUGGUACAAAGGGCAGCGAAACCGCGAGGUGAAGCCAAUCCCAUAAAGCCAUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAAGCCGGAAUUGCUAGUAAUCGCGGAUC .(((((((.....(((..((...(((....)))...))....))).....)))))))....(((((..(((((((........))))))).....))))).((((.(....).))))... ( -41.30) >Banth.0 266645 120 + 5227293/1480-1600 UGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCGCUAGUAAUCGCGGAUC .(((((((.....(((..((...(((....)))...))....))).....)))))))...(((((((((((((((........)))))).....(((((.....))))))))).))))). ( -38.30) >Bsubt.0 9591 120 + 4214630/1480-1600 UGGACAGAACAAAGGGCAGCGAAACCGCGAGGUUAAGCCAAUCCCACAAAUCUGUUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGCUGGAAUCGCUAGUAAUCGCGGAUC .(((((((.....(((..((..((((....))))..))....))).....)))))))........((((((((((........))))))((((.(((((.....)))))..)))).)))) ( -41.90) >Blich.0 9688 120 + 4222334/1480-1600 UGGGCAGAACAAAGGGCAGCGAAGCCGCGAGGCUAAGCCAAUCCCACAAAUCUGUUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGCUGGAAUCGCUAGUAAUCGCGGAUC .(((((((.....(((..((..((((....))))..))....))).....)))))))........((((((((((........))))))((((.(((((.....)))))..)))).)))) ( -43.80) >consensus UGGACGGUACAAAGGGCAGCGAAACCGCGAGGUGAAGCCAAUCCCACAAAGCCGUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAAGCUGGAAUCGCUAGUAAUCGCGGAUC .(((((((.....(((..((...(((....)))...))....))).....)))))))....(((((..(((((((........))))))).....))))).((((........))))... (-43.86 = -39.70 + -4.16)

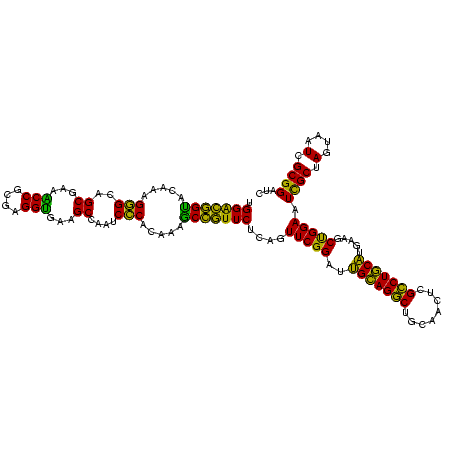
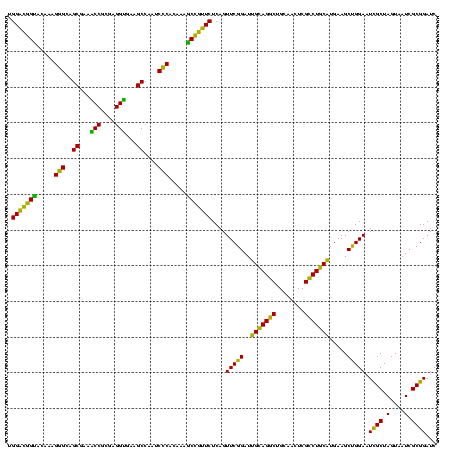
| Location | 117,184 – 117,303 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.37 |
| Mean single sequence MFE | -41.96 |
| Consensus MFE | -39.88 |
| Energy contribution | -39.72 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 119 + 4202352/1680-1800 UGGGGUAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUACGGAGUUUUUAAC .(((((.(((((((-(...((.(((......))).)).)))((((((.......))))))...)))))(((((((((....))))))))).....)))))..(((....)))........ ( -40.50) >Bclau.0 136227 118 + 4303871/1680-1800 UGAGGCAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGUUUUUA-C ...(((..((....-)).)))..(((((...)))))..((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..)).........-. ( -42.20) >Banth.0 266645 118 + 5227293/1680-1800 UGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA-- ...(((..((.....)).)))..(((((...)))))..((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..)).........-- ( -41.50) >Bsubt.0 9591 119 + 4214630/1680-1800 UGAGGUAACCUUUUAGGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUUUA-C (((((((.((((..(((((((..(((((...))))).....((((((.......)))))).....))((((((((((....))))))))))......)))))....)))).)))))))-. ( -44.00) >Blich.0 9688 118 + 4222334/1680-1800 UGAGGUAACCUUUU-GGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUA-C ...((((.((((((-(...((.(((......))).)).)))).((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))).))))...-. ( -41.60) >consensus UGAGGUAACCUUUU_GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAGAUUUUA_C ...(((..((.....)).)))..(((((...))))).......((..(((((((.((....))....((((((((((....))))))))))..))))).))..))............... (-39.88 = -39.72 + -0.16)

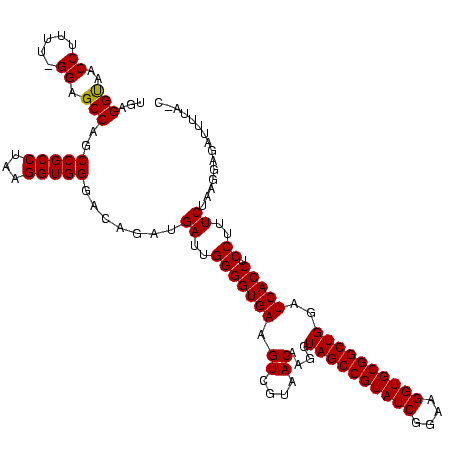
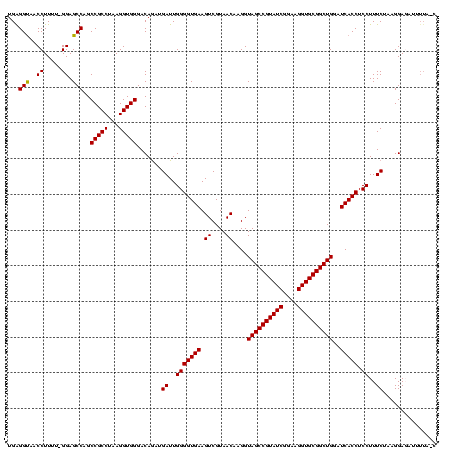
| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.99 |
| Mean single sequence MFE | -45.86 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.98 |
| Structure conservation index | 0.65 |
| SVM decision value | 4.44 |
| SVM RNA-class probability | 0.999897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/1720-1840 GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUACGGAGUUUUUAACUCUAGUCGACCAUUCGCAGAUUCAUCUGCGGAGGGACGCU ..((((((((((((..(((((..((((.(((((((((....)))))))))((....)).))))..)))))..))....)))))))))).((.((((((((....)))))))).))..... ( -50.20) >Bclau.0 136227 115 + 4303871/1720-1840 AAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGUUUUUA-CUCUAGUCGAUGUAU----GACCUUCUGGUCAUAUAGCGCU ..((((((((((((((...........((((((((((....))))))))))......((((.......)))).)))))-))))))))).(((((----((((....)))))))))..... ( -50.10) >Banth.0 266645 111 + 5227293/1720-1840 GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA----UGAACGCUGUUC-----AUCAAUAUAAGUUUCCGUGUU ...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))(((((((((((((----(((((...))))-----)))))).....)))))))).. ( -45.80) >Bsubt.0 9591 118 + 4214630/1720-1840 GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUUUA-CGGAAUAUAAGACCUUG-GGUCUUAUAAACAGAACGUUCCC .......((..((...((((((...((((((((((((....))))))))).....)))((((.....))))....)))-)))..(((((((((...-))))))))).......))..)). ( -41.30) >Blich.0 9688 119 + 4222334/1720-1840 GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUA-CGGAAUAUAAGACCUUGCGGUCUUAUAGACAGGUGCGUUUG ...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))...........((-(.(..(((((((((....)))))))))..)..)))...... ( -41.90) >consensus GAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAGAUUUUA_CGCUAGACGAGACCU_G__GACUUAUAGACAGAACCGUGCU ...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))....................................................... (-29.70 = -29.70 + -0.00)

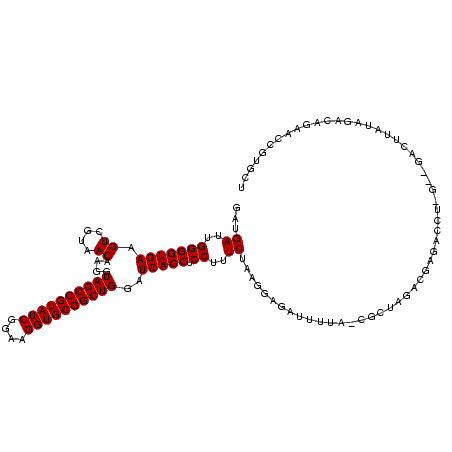
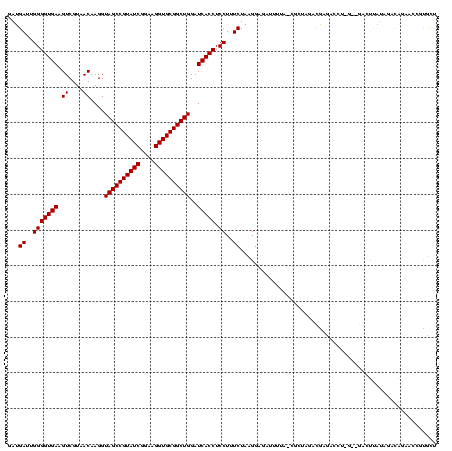
| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.99 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -15.20 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/1720-1840 AGCGUCCCUCCGCAGAUGAAUCUGCGAAUGGUCGACUAGAGUUAAAAACUCCGUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC ((((.((...((((((....))))))...)).)).)).((((.....))))(((((.((((.((....))(((((............))))).)))).)))))................. ( -32.50) >Bclau.0 136227 115 + 4303871/1720-1840 AGCGCUAUAUGACCAGAAGGUC----AUACAUCGACUAGAG-UAAAAACUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUU ((((...(((((((....))))----)))...)).)).(((-(....))))....((..((.(((((...(((((............))))).....((....)).)))))))..))... ( -27.70) >Banth.0 266645 111 + 5227293/1720-1840 AACACGGAAACUUAUAUUGAU-----GAACAGCGUUCA----UCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC .....(((.......((((((-----((((...)))))----)))))..)))...((..((.(((((...(((((............))))).....((....)).)))))))..))... ( -26.80) >Bsubt.0 9591 118 + 4214630/1720-1840 GGGAACGUUCUGUUUAUAAGACC-CAAGGUCUUAUAUUCCG-UAAAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC (((........(..(((((((((-...)))))))))..)((-(((....((((.....))))(((.....(((((............))))))))...))))).......)))....... ( -29.50) >Blich.0 9688 119 + 4222334/1720-1840 CAAACGCACCUGUCUAUAAGACCGCAAGGUCUUAUAUUCCG-UAUAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC ...........((((((((((((....)))))))))..(((-(((....((((.....))))((((.........)))).....))))))............)))............... ( -30.90) >consensus AGCACCCAACCGUAUAUAAGUC__C_AAACAUCGUAUACAG_UAAAAUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCUUGUUACGACUUCACCCCAAUCAUC .......................................................((..((.(((((...(((((............))))).....((....)).)))))))..))... (-15.20 = -15.20 + 0.00)

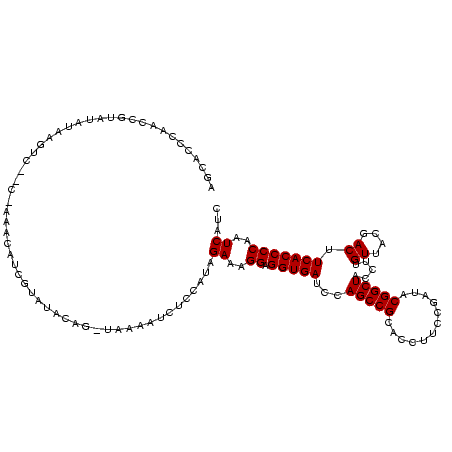
| Location | 117,184 – 117,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.60 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -8.40 |
| Energy contribution | -8.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.28 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 117184 120 + 4202352/1756-1876 AGUUUUGAGUUCUCUCAAAACUGAACAAAAGCUUCAAGCGUCCCUCCGCAGAUGAAUCUGCGAAUGGUCGACUAGAGUUAAAAACUCCGUAGAAAGGAGGUGAUCCAGCCGCACCUUCCG (((((((((....)))))))))..........(((.((((.((...((((((....))))))...)).)).)).((((.....))))....))).(((((((.........))))))).. ( -35.20) >Bclau.0 136227 114 + 4303871/1756-1876 AGUUUUGAGUCCUCUCAAAACUGAACAACAGC-ACAAGCGCUAUAUGACCAGAAGGUC----AUACAUCGACUAGAG-UAAAAACUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCG (((((((((....)))))))))((.....(((-......))).(((((((....))))----)))..))..((((((-(....))))..)))...(((((((.........))))))).. ( -32.20) >Banth.0 266645 105 + 5227293/1756-1876 AGAU---AGUUCUCUCAAAACUGAACAAAA---CGAAACACGGAAACUUAUAUUGAU-----GAACAGCGUUCA----UCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCG ....---.((((..........))))....---.......(((((......((((((-----((((...)))))----))))).((((.......))))(((.......)))...))))) ( -23.10) >Bsubt.0 9591 114 + 4214630/1756-1876 GGAACAAAGUUCCUUCAAAACUAAACAAGA----CAGGGAACGUUCUGUUUAUAAGACC-CAAGGUCUUAUAUUCCG-UAAAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCG ((((....(((((((...............----.)))))))........(((((((((-...))))))))))))).-.................(((((((.........))))))).. ( -28.09) >Blich.0 9688 115 + 4222334/1756-1876 UGAAGGAAGUUCCUUCAAAACUAAACAAGA----UCCAAACGCACCUGUCUAUAAGACCGCAAGGUCUUAUAUUCCG-UAUAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCG ((((((.....))))))...((((....((----(....(((.....(..(((((((((....)))))))))..)))-).....))).))))...(((((((.........))))))).. ( -31.10) >consensus AGAUUUAAGUUCUCUCAAAACUGAACAAAA____CAAGCACCCAACCGUAUAUAAGUC__C_AAACAUCGUAUACAG_UAAAAUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCG ...............................................................................................(((((((.........))))))).. ( -8.40 = -8.40 + 0.00)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:25:49 2006