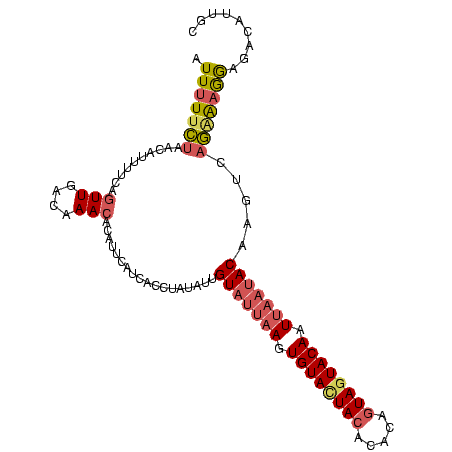
| Sequence ID | Blich.0 |
|---|---|
| Location | 3,068,831 – 3,068,932 |
| Length | 101 |
| Max. P | 0.999976 |

| Location | 3,068,831 – 3,068,932 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 62.40 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -11.86 |
| Energy contribution | -14.55 |
| Covariance contribution | 2.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.50 |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.999357 |
| Prediction | RNA |
| WARNING | Sequence 2: Base composition out of range. |
Download alignment: ClustalW | MAF
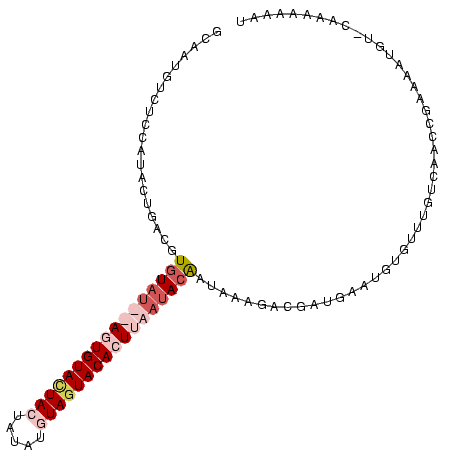
>Blich.0 3068831 101 + 4222334 UUGCUC-CUUUACAUGUGUGUGUUA--ACUGUAUUACUCGAUGUAGUACACUUAAUACAAUAUAGACAUCGUCCUUUUUUGUCAAUCGCUUUUUU-CAAAAAAAU ..((..-.......(((((((((((--(.((((((((.....)))))))).))))))).)))))((((...........))))....))......-......... ( -21.80) >Bsubt.0 3118473 103 + 4214630 UAAAUGAUUUUAUAUUAGUGUCUAU--AGUGUAUUACUUCAAUUAGUACACUUAAUACAAUAAAUACUUUGACUCACAAAGUCAAUCCUUUUUUUACAAAAAAAC ............((((.((((....--(((((((((.......)))))))))..))))))))......((((((.....)))))).................... ( -15.20) >Bcere.0 1352861 104 - 5224283 GCAAUGUCUCCUUUCUGACUUGUAUUAAGUGUACUACUAUGUGUAAUACACUUAAUACAAUAAAGGUGAUGAAUGUGUUUGUCAACUGAAAAUGU-UAGAAAAAU ...........((((((((((((((((((((((.(((.....))).))))))))))))))...((.(((..((....))..))).))......))-))))))... ( -31.20) >Banth.0 1299781 104 - 5227293 GCAAUGUCUCCUUCCUGACAUGUAUUAAUUGUACUAAUGUGUGUAGUACACUUAAUACGAUAUAGGUGAUAAAUGUGUUUGUCAACUGAAAAUGU-UAGAAAAAU ..............((((((((((((((.(((((((.......))))))).)))))))....(((.(((((((....))))))).)))...))))-)))...... ( -26.70) >consensus GCAAUGUCUCCAUACUGACGUGUAU__AGUGUACUACUAUAUGUAGUACACUUAAUACAAUAAAGACGAUGAAUGUGUUUGUCAACCGAAAAUGU_CAAAAAAAU ....................(((((((((((((((((.....))))))))))))))))).............................................. (-11.86 = -14.55 + 2.69)

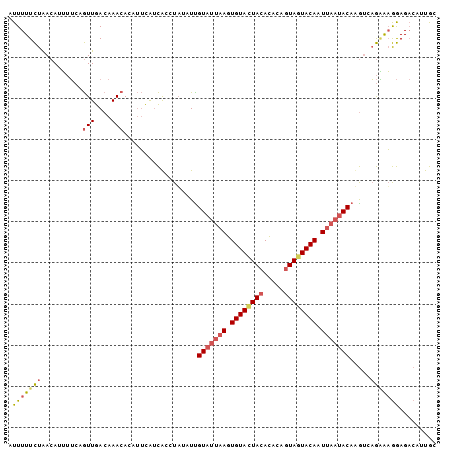
| Location | 3,068,831 – 3,068,932 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 62.40 |
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -10.30 |
| Energy contribution | -13.43 |
| Covariance contribution | 3.12 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.995969 |
| Prediction | RNA |
| WARNING | Sequence 2: Base composition out of range. |
Download alignment: ClustalW | MAF
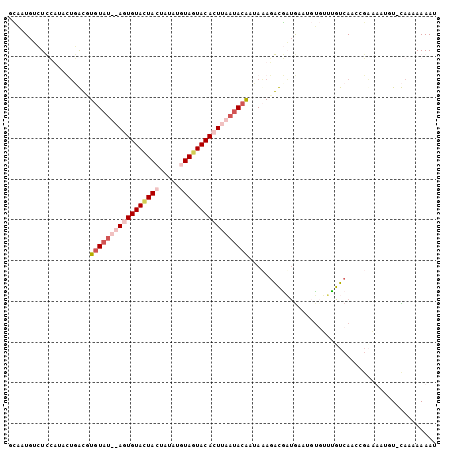
>Blich.0 3068831 101 + 4222334 AUUUUUUUG-AAAAAAGCGAUUGACAAAAAAGGACGAUGUCUAUAUUGUAUUAAGUGUACUACAUCGAGUAAUACAGU--UAACACACACAUGUAAAG-GAGCAA .........-......((..((.(((.....((((...))))....(((.((((.((((.(((.....))).)))).)--))).)))....))).)).-..)).. ( -15.10) >Bsubt.0 3118473 103 + 4214630 GUUUUUUUGUAAAAAAAGGAUUGACUUUGUGAGUCAAAGUAUUUAUUGUAUUAAGUGUACUAAUUGAAGUAAUACACU--AUAGACACUAAUAUAAAAUCAUUUA ..((((((....))))))((((.((((((.....)))))).....((((((((.((((.(((..((.(....).))..--.)))))))))))))))))))..... ( -16.80) >Bcere.0 1352861 104 - 5224283 AUUUUUCUA-ACAUUUUCAGUUGACAAACACAUUCAUCACCUUUAUUGUAUUAAGUGUAUUACACAUAGUAGUACACUUAAUACAAGUCAGAAAGGAGACAUUGC ...(((((.-((.......(((....)))................((((((((((((((((((.....)))))))))))))))))))).)))))(....)..... ( -29.70) >Banth.0 1299781 104 - 5227293 AUUUUUCUA-ACAUUUUCAGUUGACAAACACAUUUAUCACCUAUAUCGUAUUAAGUGUACUACACACAUUAGUACAAUUAAUACAUGUCAGGAAGGAGACAUUGC ..((((((.-.........(((....)))..........(((((((.(((((((.(((((((.......))))))).))))))))))).))).))))))...... ( -21.50) >consensus AUUUUUCUA_AAAAAAUCAAUUGACAAACACAGUCAACACCUAUAUUGUAUUAAGUGUACUACACAAAGUAAUACACU__AUACACACCAAUAAAAAGACAUUGA ....((((...........((((((.......)))))).........((((((((((((((((.....))))))))))))))))..........))))....... (-10.30 = -13.43 + 3.12)

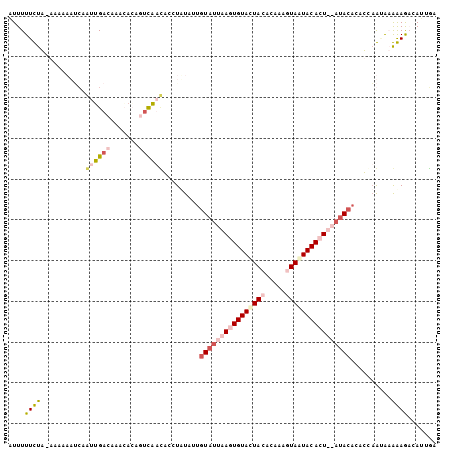
| Location | 3,068,831 – 3,068,932 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 66.67 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -13.46 |
| Energy contribution | -16.03 |
| Covariance contribution | 2.57 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.51 |
| SVM decision value | 5.14 |
| SVM RNA-class probability | 0.999976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
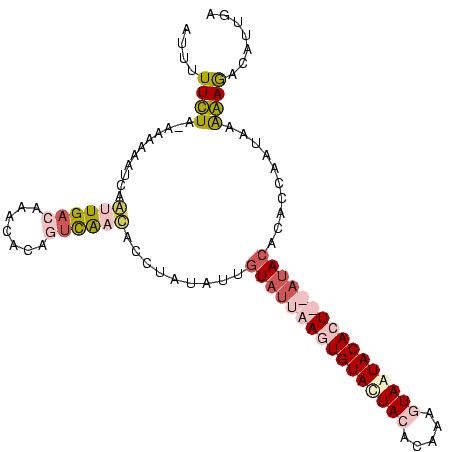
>Blich.0 3068831 101 + 4222334/0-105 UUGCUC-CUUUACAUGUGUGUGUUA--ACUGUAUUACUCGAUGUAGUACACUUAAUACAAUAUAGACAUCGUCCUUUUUUGUCAAUCGCUUUUUUCAAAAAAAU ..((..-.......(((((((((((--(.((((((((.....)))))))).))))))).)))))((((...........))))....))............... ( -21.80) >Bcere.0 1352861 104 - 5224283/0-105 GCAAUGUCUCCUUUCUGACUUGUAUUAAGUGUACUACUAUGUGUAAUACACUUAAUACAAUAAAGGUGAUGAAUGUGUUUGUCAACUGAAAAUGUUAGAAAAAU ...........((((((((((((((((((((((.(((.....))).))))))))))))))...((.(((..((....))..))).))......))))))))... ( -31.20) >Banth.0 1299781 104 - 5227293/0-105 GCAAUGUCUCCUUCCUGACAUGUAUUAAUUGUACUAAUGUGUGUAGUACACUUAAUACGAUAUAGGUGAUAAAUGUGUUUGUCAACUGAAAAUGUUAGAAAAAU ..............((((((((((((((.(((((((.......))))))).)))))))....(((.(((((((....))))))).)))...)))))))...... ( -26.70) >consensus GCAAUGUCUCCUUACUGACAUGUAUUAACUGUACUACUAUGUGUAGUACACUUAAUACAAUAUAGGUGAUGAAUGUGUUUGUCAACUGAAAAUGUUAGAAAAAU ..............(((((.((((((((.((((((((.....)))))))).)))))))).....(((((((((....)))))).)))......)))))...... (-13.46 = -16.03 + 2.57)

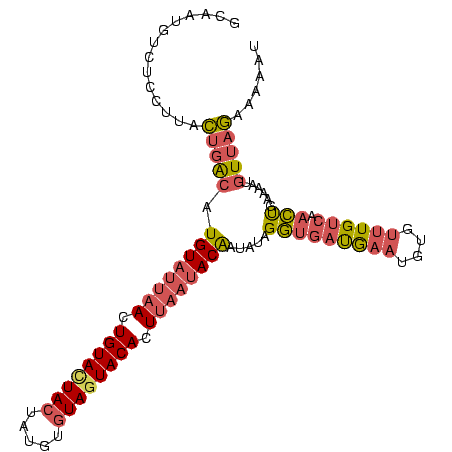

| Location | 3,068,831 – 3,068,932 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 66.67 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -11.02 |
| Energy contribution | -13.03 |
| Covariance contribution | 2.01 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.50 |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 3068831 101 + 4222334/0-105 AUUUUUUUGAAAAAAGCGAUUGACAAAAAAGGACGAUGUCUAUAUUGUAUUAAGUGUACUACAUCGAGUAAUACAGU--UAACACACACAUGUAAAG-GAGCAA ...............((..((.(((.....((((...))))....(((.((((.((((.(((.....))).)))).)--))).)))....))).)).-..)).. ( -15.10) >Bcere.0 1352861 104 - 5224283/0-105 AUUUUUCUAACAUUUUCAGUUGACAAACACAUUCAUCACCUUUAUUGUAUUAAGUGUAUUACACAUAGUAGUACACUUAAUACAAGUCAGAAAGGAGACAUUGC ...(((((.((.......(((....)))................((((((((((((((((((.....)))))))))))))))))))).)))))(....)..... ( -29.70) >Banth.0 1299781 104 - 5227293/0-105 AUUUUUCUAACAUUUUCAGUUGACAAACACAUUUAUCACCUAUAUCGUAUUAAGUGUACUACACACAUUAGUACAAUUAAUACAUGUCAGGAAGGAGACAUUGC ..((((((..........(((....)))..........(((((((.(((((((.(((((((.......))))))).))))))))))).))).))))))...... ( -21.50) >consensus AUUUUUCUAACAUUUUCAGUUGACAAACACAUUCAUCACCUAUAUUGUAUUAAGUGUACUACACACAGUAGUACAAUUAAUACAAGUCAGAAAGGAGACAUUGC .(((((((..........(((....)))..................(((((((.((((((((.....)))))))).))))))).....)))))))......... (-11.02 = -13.03 + 2.01)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:25:23 2006