| Sequence ID | Bcere.0 |
|---|---|
| Location | 3,365,488 – 4,806,405 |
| Length | 1440917 |
| Max. P | 0.999998 |

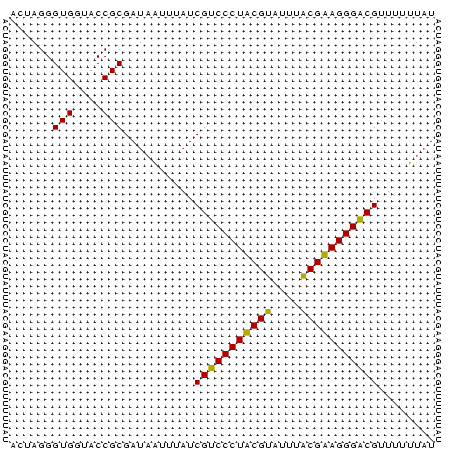
| Location | 4,806,285 – 4,806,405 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.33 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -18.54 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.63 |
| SVM decision value | 6.40 |
| SVM RNA-class probability | 0.999998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
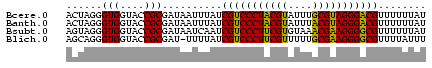
>Bcere.0 4806285 120 + 5224283/8-128 GGUACUAAAAAGUGCCUAUGUUACUUUUUGGUACCUAUCGUACUUUUAAGUGCGUUCUUCAAAAAAAGAGCGAAGCCAUUCAUAAUAGCAUAUAUGAGCAAUAAAGUUAUUUUUGUAGAA ((((((((((((((.......))))))))))))))...((((((....))))))((((.((((((...(((.......((((((........)))))).......))).)))))).)))) ( -31.64) >Banth.0 4814425 119 + 5227293/8-128 GGUACUGAAAAGUGACUAUGUUACUUUUUGGUGCCUAUCGUACUUUAAAGCGCGUUCUUCAAAAAAAGAAUGAAACCAUUCAUAAUAGCAUUUGUAGAGAUUAAAUAUAUUUUU-UAGAA (((((..(((((((((...)))))))))..)))))...((..((....))..))....((.(((((((((((....))))).((((..(.......)..))))......)))))-).)). ( -27.70) >Blich.0 2724962 120 + 4222334/8-128 GGUACCGAAAAGUACCUAUCUCACUUUUUGGUUCCUAUCACACUUAAAAGUGCGUACUUUUUUUCGCAGAUGAUAUGUACCAUAAUAUCAUUAGCCGGCAGCCAUUUGCUAUACCUUAAA (((((((((((((((......(((((((.(((.........))).))))))).))))))))....((.((((((((........)))))))).))))).(((.....))).))))..... ( -29.10) >consensus GGUACUGAAAAGUGCCUAUGUUACUUUUUGGUACCUAUCGUACUUUAAAGUGCGUUCUUCAAAAAAAGAAUGAAACCAUUCAUAAUAGCAUUUGUAGGCAAUAAAUUUAUUUUU_UAGAA ((((((((((((((.......)))))))))))))).................(((((((......)))))))................................................ (-18.54 = -19.00 + 0.46)

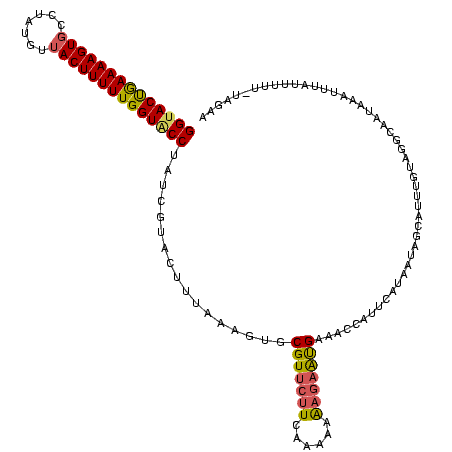
| Location | 3,365,488 – 3,365,548 |
|---|---|
| Length | 60 |
| Sequences | 4 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -18.78 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.43 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 3365488 60 - 5224283 AUAAAAAACGUCCCUACGCAAAUACGUAGGGACGAUAAAUUAUCGCGGUACCACCCUAGU ........((((((((((......))))))))))............((.....))..... ( -19.60) >Banth.0 3505951 60 - 5227293 AUAAAAAACGUCCCUACGUAAAUACGUAGGGACGAUAAAUUAUCGCGGUACCACCCUAGU ........(((((((((((....)))))))))))............((.....))..... ( -21.00) >Bsubt.0 3036937 60 + 4214630 AUAAAAAACGCCCCUUCGUUUACACGAAGGGACGAUUGAUUAUCGCGGUACCACCCUACU ........(((((((((((....))))))))..(((.....))))))............. ( -17.30) >Blich.0 3000788 59 + 4222334 AAAUAAAACGCCCCUUCGCAAAAACGAAGGGACGAUAAAA-AUCGCGGUACCACCCUGCU ........((.(((((((......))))))).))......-...((((.......)))). ( -17.20) >consensus AUAAAAAACGCCCCUACGCAAAUACGAAGGGACGAUAAAUUAUCGCGGUACCACCCUACU ........((((((((((......))))))))))............((.....))..... (-17.60 = -17.60 + -0.00)
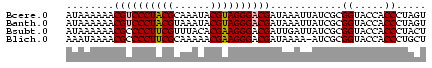
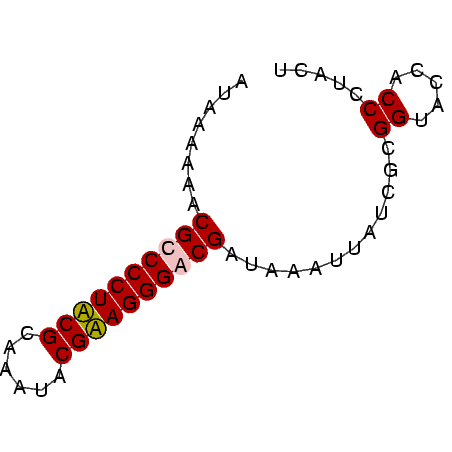
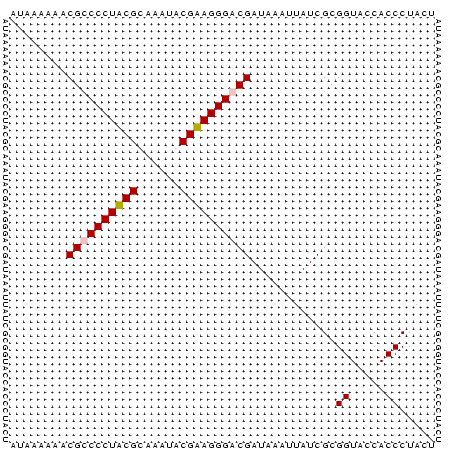
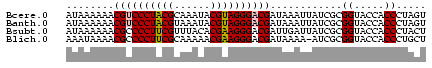
| Location | 3,365,488 – 3,365,548 |
|---|---|
| Length | 60 |
| Sequences | 4 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -23.35 |
| Energy contribution | -22.35 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -4.10 |
| Structure conservation index | 1.02 |
| SVM decision value | 3.44 |
| SVM RNA-class probability | 0.999214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 3365488 60 - 5224283 ACUAGGGUGGUACCGCGAUAAUUUAUCGUCCCUACGUAUUUGCGUAGGGACGUUUUUUAU ......(((....))).((((.....(((((((((((....)))))))))))....)))) ( -22.90) >Banth.0 3505951 60 - 5227293 ACUAGGGUGGUACCGCGAUAAUUUAUCGUCCCUACGUAUUUACGUAGGGACGUUUUUUAU ......(((....))).((((.....(((((((((((....)))))))))))....)))) ( -22.60) >Bsubt.0 3036937 60 + 4214630 AGUAGGGUGGUACCGCGAUAAUCAAUCGUCCCUUCGUGUAAACGAAGGGGCGUUUUUUAU ......(((....))).((((.....(((((((((((....)))))))))))....)))) ( -22.70) >Blich.0 3000788 59 + 4222334 AGCAGGGUGGUACCGCGAU-UUUUAUCGUCCCUUCGUUUUUGCGAAGGGGCGUUUUAUUU .((.((......))))...-......(((((((((((....)))))))))))........ ( -23.40) >consensus ACUAGGGUGGUACCGCGAUAAUUUAUCGUCCCUACGUAUUUACGAAGGGACGUUUUUUAU ......(((....)))..........(((((((((((....)))))))))))........ (-23.35 = -22.35 + -1.00)
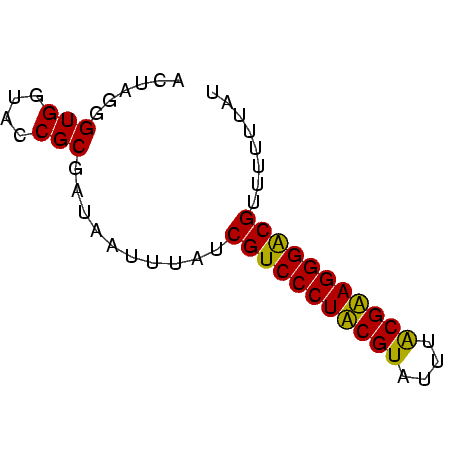
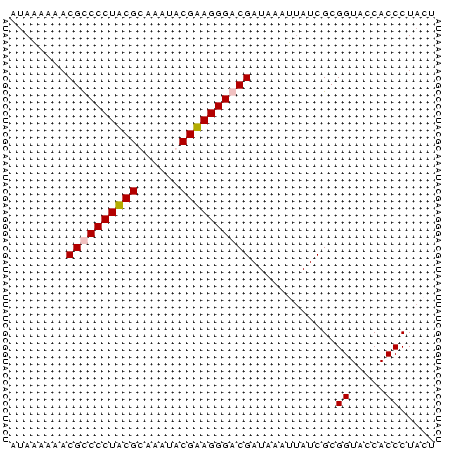
| Location | 3,365,488 – 3,365,548 |
|---|---|
| Length | 60 |
| Sequences | 4 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -18.78 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.43 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 3365488 60 - 5224283/0-60 AUAAAAAACGUCCCUACGCAAAUACGUAGGGACGAUAAAUUAUCGCGGUACCACCCUAGU ........((((((((((......))))))))))............((.....))..... ( -19.60) >Banth.0 3505951 60 - 5227293/0-60 AUAAAAAACGUCCCUACGUAAAUACGUAGGGACGAUAAAUUAUCGCGGUACCACCCUAGU ........(((((((((((....)))))))))))............((.....))..... ( -21.00) >Bsubt.0 3036937 60 + 4214630/0-60 AUAAAAAACGCCCCUUCGUUUACACGAAGGGACGAUUGAUUAUCGCGGUACCACCCUACU ........(((((((((((....))))))))..(((.....))))))............. ( -17.30) >Blich.0 3000788 59 + 4222334/0-60 AAAUAAAACGCCCCUUCGCAAAAACGAAGGGACGAUAAAA-AUCGCGGUACCACCCUGCU ........((.(((((((......))))))).))......-...((((.......)))). ( -17.20) >consensus AUAAAAAACGCCCCUACGCAAAUACGAAGGGACGAUAAAUUAUCGCGGUACCACCCUACU ........((((((((((......))))))))))............((.....))..... (-17.60 = -17.60 + -0.00)

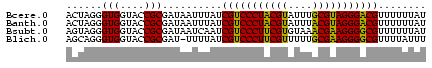
| Location | 3,365,488 – 3,365,548 |
|---|---|
| Length | 60 |
| Sequences | 4 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -23.35 |
| Energy contribution | -22.35 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -4.10 |
| Structure conservation index | 1.02 |
| SVM decision value | 3.44 |
| SVM RNA-class probability | 0.999214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 3365488 60 - 5224283/0-60 ACUAGGGUGGUACCGCGAUAAUUUAUCGUCCCUACGUAUUUGCGUAGGGACGUUUUUUAU ......(((....))).((((.....(((((((((((....)))))))))))....)))) ( -22.90) >Banth.0 3505951 60 - 5227293/0-60 ACUAGGGUGGUACCGCGAUAAUUUAUCGUCCCUACGUAUUUACGUAGGGACGUUUUUUAU ......(((....))).((((.....(((((((((((....)))))))))))....)))) ( -22.60) >Bsubt.0 3036937 60 + 4214630/0-60 AGUAGGGUGGUACCGCGAUAAUCAAUCGUCCCUUCGUGUAAACGAAGGGGCGUUUUUUAU ......(((....))).((((.....(((((((((((....)))))))))))....)))) ( -22.70) >Blich.0 3000788 59 + 4222334/0-60 AGCAGGGUGGUACCGCGAU-UUUUAUCGUCCCUUCGUUUUUGCGAAGGGGCGUUUUAUUU .((.((......))))...-......(((((((((((....)))))))))))........ ( -23.40) >consensus ACUAGGGUGGUACCGCGAUAAUUUAUCGUCCCUACGUAUUUACGAAGGGACGUUUUUUAU ......(((....)))..........(((((((((((....)))))))))))........ (-23.35 = -22.35 + -1.00)
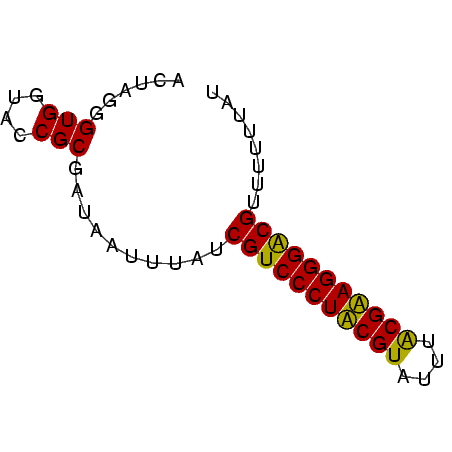
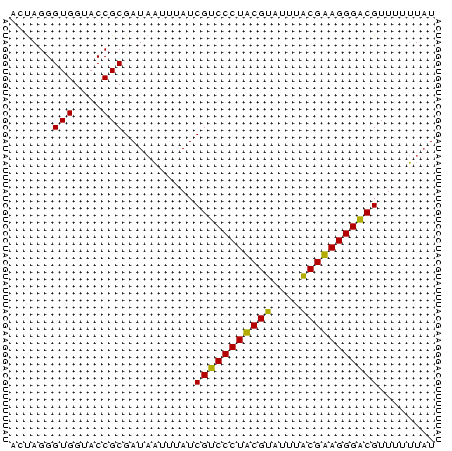
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:25:19 2006