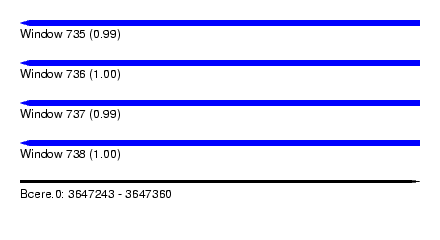
| Sequence ID | Bcere.0 |
|---|---|
| Location | 3,647,243 – 3,647,360 |
| Length | 117 |
| Max. P | 0.998780 |

| Location | 3,647,243 – 3,647,360 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.57 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -17.31 |
| Energy contribution | -17.76 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.15 |
| Mean z-score | -4.57 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 3647243 117 - 5224283 CUCUCUCCUUUUCCAUCGAUCAUCUCAUGGAAAGUGGAAAGGUUCUGCUAUGCCUACUUUUUCAUAGAACGAAAAAACCCUUUGUGUUUUACACAAAGGGAGAAUUUA-AGGCACAUC .(((.(((((((((((.((.....)))))))))).))).)))........(((((..((((((.......)))))).(((((((((.....)))))))))........-))))).... ( -36.70) >Banth.0 3804432 116 - 5227293 CUCUCUCCUUUUCCAUCGAUCAUCUCAUGGAAAGUGGAAAGGUUCUGCUAUGCCUA-UUUUUCAUAGAACGAAAAAACCCUUUGUGUUUUACACAAAGGGAGAAUUUA-AGGCACAUC .(((.(((((((((((.((.....)))))))))).))).)))........(((((.-((((((.......)))))).(((((((((.....)))))))))........-))))).... ( -36.90) >Blich.0 2381057 107 + 4222334 CUCUCUCCUUUUCGAUCA--CAUCUCACGAAAAGAGGAAUGGUUCU--UUCCCCUG-----UCCUAAAC--AAAAAACCCGCUUUAUUGAAAAAGCGGGGCUGUUUUACAGACAGGUC ...((..(((((((....--.......)))))))..))..((....--..))((((-----((..((((--(.....((((((((......))))))))..)))))....)))))).. ( -32.90) >consensus CUCUCUCCUUUUCCAUCGAUCAUCUCAUGGAAAGUGGAAAGGUUCUGCUAUGCCUA_UUUUUCAUAGAACGAAAAAACCCUUUGUGUUUUACACAAAGGGAGAAUUUA_AGGCACAUC ...((..((((((...............))))))..))..(((........))).......................(((((((((.....))))))))).................. (-17.31 = -17.76 + 0.45)

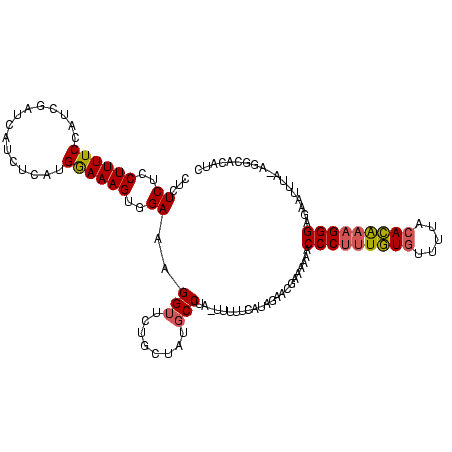
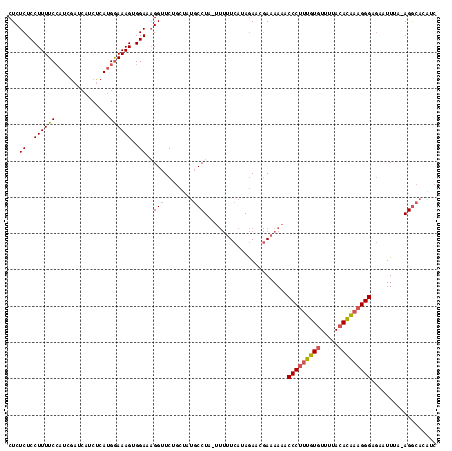
| Location | 3,647,243 – 3,647,360 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.57 |
| Mean single sequence MFE | -40.03 |
| Consensus MFE | -21.14 |
| Energy contribution | -24.60 |
| Covariance contribution | 3.46 |
| Combinations/Pair | 1.16 |
| Mean z-score | -4.49 |
| Structure conservation index | 0.53 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 3647243 117 - 5224283 GAUGUGCCU-UAAAUUCUCCCUUUGUGUAAAACACAAAGGGUUUUUUCGUUCUAUGAAAAAGUAGGCAUAGCAGAACCUUUCCACUUUCCAUGAGAUGAUCGAUGGAAAAGGAGAGAG ..(((((((-........(((((((((.....)))))))))(((((((((...))))))))).))))))).......((((((..(((((((((.....)).))))))).)))))).. ( -41.50) >Banth.0 3804432 116 - 5227293 GAUGUGCCU-UAAAUUCUCCCUUUGUGUAAAACACAAAGGGUUUUUUCGUUCUAUGAAAAA-UAGGCAUAGCAGAACCUUUCCACUUUCCAUGAGAUGAUCGAUGGAAAAGGAGAGAG ..(((((((-........(((((((((.....))))))))).((((((((...))))))))-.))))))).......((((((..(((((((((.....)).))))))).)))))).. ( -40.90) >Blich.0 2381057 107 + 4222334 GACCUGUCUGUAAAACAGCCCCGCUUUUUCAAUAAAGCGGGUUUUUU--GUUUAGGA-----CAGGGGAA--AGAACCAUUCCUCUUUUCGUGAGAUG--UGAUCGAAAAGGAGAGAG ..(((((((...((((((.((((((((......))))))))....))--)))).)))-----))))((..--....)).(((..(((((((.......--....)))))))..))).. ( -37.70) >consensus GAUGUGCCU_UAAAUUCUCCCUUUGUGUAAAACACAAAGGGUUUUUUCGUUCUAUGAAAAA_UAGGCAUAGCAGAACCUUUCCACUUUCCAUGAGAUGAUCGAUGGAAAAGGAGAGAG ..(((((((.........(((((((((.....)))))))))..((((((.....))))))...))))))).......((((((..(((((((((.....)).))))))).)))))).. (-21.14 = -24.60 + 3.46)

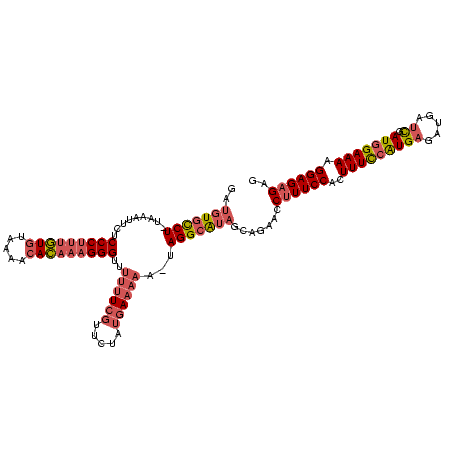
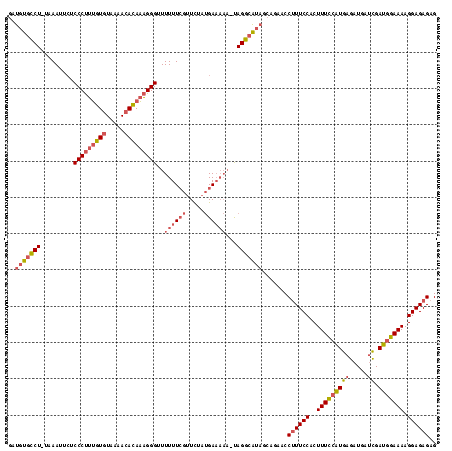
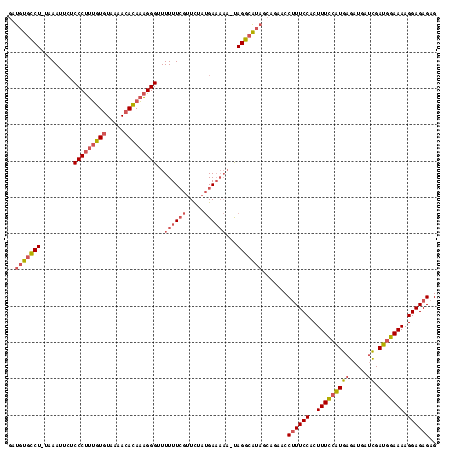
| Location | 3,647,243 – 3,647,360 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.57 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -17.31 |
| Energy contribution | -17.76 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.15 |
| Mean z-score | -4.57 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 3647243 117 - 5224283/0-118 CUCUCUCCUUUUCCAUCGAUCAUCUCAUGGAAAGUGGAAAGGUUCUGCUAUGCCUACUUUUUCAUAGAACGAAAAAACCCUUUGUGUUUUACACAAAGGGAGAAUUUA-AGGCACAUC .(((.(((((((((((.((.....)))))))))).))).)))........(((((..((((((.......)))))).(((((((((.....)))))))))........-))))).... ( -36.70) >Banth.0 3804432 116 - 5227293/0-118 CUCUCUCCUUUUCCAUCGAUCAUCUCAUGGAAAGUGGAAAGGUUCUGCUAUGCCUA-UUUUUCAUAGAACGAAAAAACCCUUUGUGUUUUACACAAAGGGAGAAUUUA-AGGCACAUC .(((.(((((((((((.((.....)))))))))).))).)))........(((((.-((((((.......)))))).(((((((((.....)))))))))........-))))).... ( -36.90) >Blich.0 2381057 107 + 4222334/0-118 CUCUCUCCUUUUCGAUCA--CAUCUCACGAAAAGAGGAAUGGUUCU--UUCCCCUG-----UCCUAAAC--AAAAAACCCGCUUUAUUGAAAAAGCGGGGCUGUUUUACAGACAGGUC ...((..(((((((....--.......)))))))..))..((....--..))((((-----((..((((--(.....((((((((......))))))))..)))))....)))))).. ( -32.90) >consensus CUCUCUCCUUUUCCAUCGAUCAUCUCAUGGAAAGUGGAAAGGUUCUGCUAUGCCUA_UUUUUCAUAGAACGAAAAAACCCUUUGUGUUUUACACAAAGGGAGAAUUUA_AGGCACAUC ...((..((((((...............))))))..))..(((........))).......................(((((((((.....))))))))).................. (-17.31 = -17.76 + 0.45)

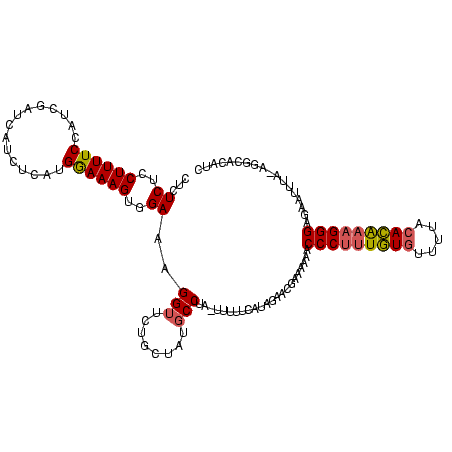

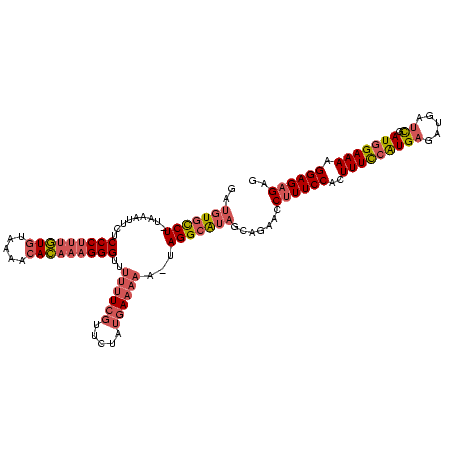
| Location | 3,647,243 – 3,647,360 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.57 |
| Mean single sequence MFE | -40.03 |
| Consensus MFE | -21.14 |
| Energy contribution | -24.60 |
| Covariance contribution | 3.46 |
| Combinations/Pair | 1.16 |
| Mean z-score | -4.49 |
| Structure conservation index | 0.53 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 3647243 117 - 5224283/0-118 GAUGUGCCU-UAAAUUCUCCCUUUGUGUAAAACACAAAGGGUUUUUUCGUUCUAUGAAAAAGUAGGCAUAGCAGAACCUUUCCACUUUCCAUGAGAUGAUCGAUGGAAAAGGAGAGAG ..(((((((-........(((((((((.....)))))))))(((((((((...))))))))).))))))).......((((((..(((((((((.....)).))))))).)))))).. ( -41.50) >Banth.0 3804432 116 - 5227293/0-118 GAUGUGCCU-UAAAUUCUCCCUUUGUGUAAAACACAAAGGGUUUUUUCGUUCUAUGAAAAA-UAGGCAUAGCAGAACCUUUCCACUUUCCAUGAGAUGAUCGAUGGAAAAGGAGAGAG ..(((((((-........(((((((((.....))))))))).((((((((...))))))))-.))))))).......((((((..(((((((((.....)).))))))).)))))).. ( -40.90) >Blich.0 2381057 107 + 4222334/0-118 GACCUGUCUGUAAAACAGCCCCGCUUUUUCAAUAAAGCGGGUUUUUU--GUUUAGGA-----CAGGGGAA--AGAACCAUUCCUCUUUUCGUGAGAUG--UGAUCGAAAAGGAGAGAG ..(((((((...((((((.((((((((......))))))))....))--)))).)))-----))))((..--....)).(((..(((((((.......--....)))))))..))).. ( -37.70) >consensus GAUGUGCCU_UAAAUUCUCCCUUUGUGUAAAACACAAAGGGUUUUUUCGUUCUAUGAAAAA_UAGGCAUAGCAGAACCUUUCCACUUUCCAUGAGAUGAUCGAUGGAAAAGGAGAGAG ..(((((((.........(((((((((.....)))))))))..((((((.....))))))...))))))).......((((((..(((((((((.....)).))))))).)))))).. (-21.14 = -24.60 + 3.46)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:25:15 2006