
| Sequence ID | Bcere.0 |
|---|---|
| Location | 3,798,782 – 3,798,894 |
| Length | 112 |
| Max. P | 0.988988 |

| Location | 3,798,782 – 3,798,894 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -23.86 |
| Energy contribution | -22.36 |
| Covariance contribution | -1.49 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.595786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
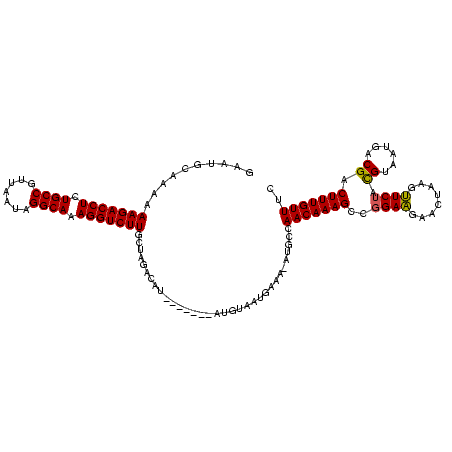
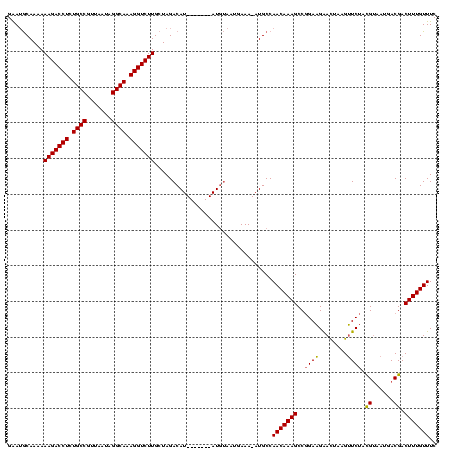
>Bcere.0 3798782 112 - 5224283/0-120 CCAAAGGGGAGUAGCGUCCAAGAAUAUUUCUUGGAAACAAAGUCGUCAUUACGUAGAACUUAGUUCUUCCGGCUUUGUUGGCAU-UUUCAUUACAU-------AUGUCUAGCAAGACCUU ......(..(((.((.(((((((.....)))))))((((((((((.........(((((...)))))..)))))))))).))))-)..).......-------..((((....))))... ( -29.10) >Banth.0 3958133 112 - 5227293/0-120 CCAAAGGGGAGUAGCGUCCAAGAAUAUUUCUUGGAAACAAAGUCGUCAUUACGUAGAACUUAGUUCUUCCGGCUUUGUUGGCAU-UUUCAUUACAU-------AUGUCUAGCAAGACCUU ......(..(((.((.(((((((.....)))))))((((((((((.........(((((...)))))..)))))))))).))))-)..).......-------..((((....))))... ( -29.10) >Blich.0 1470567 118 + 4222334/0-120 CCAAAGGGGAGUAGCGUCCGGAUGU-UUAACCGGAAACAAAGUCGUCAGUUCAUGGAUCUUCAUCCAUC-GGCUUUGUUGGCAUGUUUUAAUUAAUGAUAACCAUGUCUAGCAAGACCUU ....((((.....((.(((((....-....)))))((((((((((.......((((((....)))))))-)))))))))((((((.((...........)).))))))..))....)))) ( -31.61) >consensus CCAAAGGGGAGUAGCGUCCAAGAAUAUUUCUUGGAAACAAAGUCGUCAUUACGUAGAACUUAGUUCUUCCGGCUUUGUUGGCAU_UUUCAUUACAU_______AUGUCUAGCAAGACCUU ....((((.....((.((((((((...))))))))((((((((((......)...(((........))).)))))))))((((((.................))))))..))....)))) (-23.86 = -22.36 + -1.49)

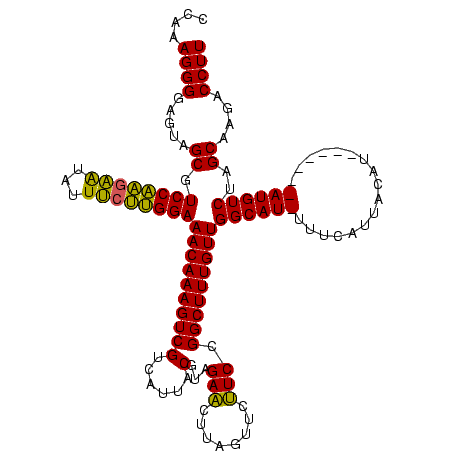
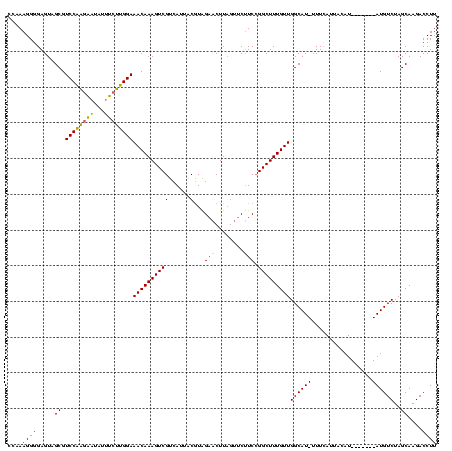
| Location | 3,798,782 – 3,798,894 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -31.06 |
| Energy contribution | -30.56 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.47 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 3798782 112 - 5224283/33-153 GAAACAAAGUCGUCAUUACGUAGAACUUAGUUCUUCCGGCUUUGUUGGCAU-UUUCAUUACAU-------AUGUCUAGCAAGACCUUUGCCUAUUAACGGCAGAGGUCUUUUUUGCAUCC ..((((((((((.........(((((...)))))..))))))))))(((((-...........-------)))))..((((((((((((((.......)))))))))))....))).... ( -36.00) >Banth.0 3958133 112 - 5227293/33-153 GAAACAAAGUCGUCAUUACGUAGAACUUAGUUCUUCCGGCUUUGUUGGCAU-UUUCAUUACAU-------AUGUCUAGCAAGACCUUUGCCUAUUAACGGCAGAGGUCUUUUUUGCAUUC ..((((((((((.........(((((...)))))..))))))))))(((((-...........-------)))))..((((((((((((((.......)))))))))))....))).... ( -36.00) >Blich.0 1470567 119 + 4222334/33-153 GAAACAAAGUCGUCAGUUCAUGGAUCUUCAUCCAUC-GGCUUUGUUGGCAUGUUUUAAUUAAUGAUAACCAUGUCUAGCAAGACCUUUGCCUAAUGUUGGCAGAGGUCUUUUUUGUUUUC ..((((((((((.......((((((....)))))))-)))))))))((((((.((...........)).))))))....((((((((((((.......)))))))))))).......... ( -39.61) >consensus GAAACAAAGUCGUCAUUACGUAGAACUUAGUUCUUCCGGCUUUGUUGGCAU_UUUCAUUACAU_______AUGUCUAGCAAGACCUUUGCCUAUUAACGGCAGAGGUCUUUUUUGCAUUC ..((((((((((......)...(((........))).)))))))))((((((.................))))))..((((((((((((((.......)))))))))))....))).... (-31.06 = -30.56 + -0.50)

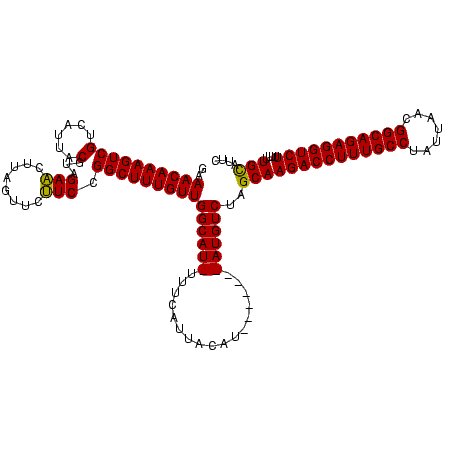
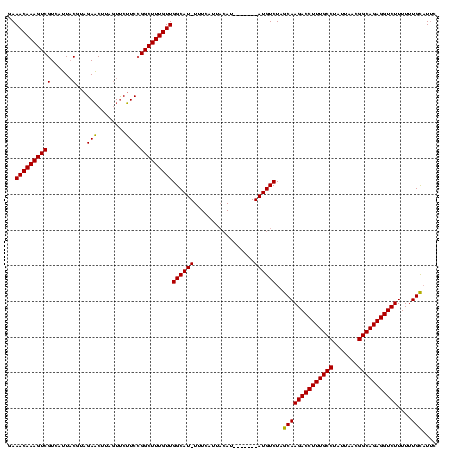
| Location | 3,798,782 – 3,798,894 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -21.56 |
| Energy contribution | -21.23 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 3798782 112 - 5224283/33-153 GGAUGCAAAAAAGACCUCUGCCGUUAAUAGGCAAAGGUCUUGCUAGACAU-------AUGUAAUGAAA-AUGCCAACAAAGCCGGAAGAACUAAGUUCUACGUAAUGACGACUUUGUUUC ((..(((....((((((.((((.......)))).)))))))))....(((-------.....)))...-...))(((((((.....(((((...))))).(((....))).))))))).. ( -27.90) >Banth.0 3958133 112 - 5227293/33-153 GAAUGCAAAAAAGACCUCUGCCGUUAAUAGGCAAAGGUCUUGCUAGACAU-------AUGUAAUGAAA-AUGCCAACAAAGCCGGAAGAACUAAGUUCUACGUAAUGACGACUUUGUUUC ....(((...(((((((.((((.......)))).)))))))......(((-------.....)))...-.))).(((((((.....(((((...))))).(((....))).))))))).. ( -27.80) >Blich.0 1470567 119 + 4222334/33-153 GAAAACAAAAAAGACCUCUGCCAACAUUAGGCAAAGGUCUUGCUAGACAUGGUUAUCAUUAAUUAAAACAUGCCAACAAAGCC-GAUGGAUGAAGAUCCAUGAACUGACGACUUUGUUUC ..........(((((((.((((.......)))).)))))))...(((((.(((..(((.............((.......)).-.((((((....))))))....)))..))).))))). ( -30.20) >consensus GAAUGCAAAAAAGACCUCUGCCGUUAAUAGGCAAAGGUCUUGCUAGACAU_______AUGUAAUGAAA_AUGCCAACAAAGCCGGAAGAACUAAGUUCUACGUAAUGACGACUUUGUUUC ..........(((((((.((((.......)))).))))))).................................(((((((..((((........)))).((......)).))))))).. (-21.56 = -21.23 + -0.33)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:25:05 2006