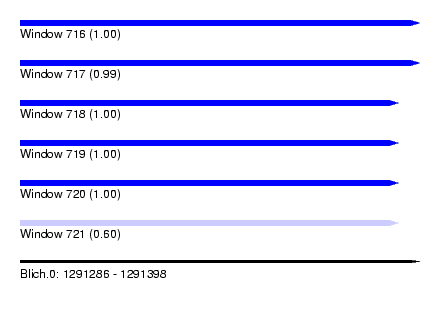
| Sequence ID | Blich.0 |
|---|---|
| Location | 1,291,286 – 1,291,398 |
| Length | 112 |
| Max. P | 0.999979 |

| Location | 1,291,286 – 1,291,398 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.11 |
| Mean single sequence MFE | -40.55 |
| Consensus MFE | -32.46 |
| Energy contribution | -33.02 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.80 |
| SVM decision value | 5.21 |
| SVM RNA-class probability | 0.999979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 1291286 112 + 4222334/0-120 CUUAUCACGAGAGGUGGAGGGACUGGCCCUUUGAAACCUCAGCAACCGGUCUGCACUGACGACGUCAGUGCACCAAGGUGCUAAAUCCAGCAAGCGG-----AUGCUU---GGAAGAUAA ..((((....(((((.(((((.....)))))....)))))((((.((....(((((((((...)))))))))....))))))...(((((((.....-----.))).)---))).)))). ( -44.10) >Bsubt.0 1257615 116 + 4214630/0-120 CUUAUCGUGAGAGGUGGAGGGACUGGCCCUUAGAAACCUCAGCAACCGGCUUGUUUUG-CAUUUGCAAAGCGCCAAGGUGCUAAAUCCAGCAAGCGUUUUUUAUGCUU---GGAAGAUAA ..(((((.(((((((.(((((.....)))))....)))))((((.(((((..((((((-(....))))))))))..))))))...)))..(((((((.....))))))---)...)))). ( -43.10) >Banth.0 2459361 111 + 5227293/0-120 CUUAUCCAGAGAGGUGGAGGGAACGGCCCUAUGAAACCUCAGCAACCCCUAUGUAAAU---------GCAUAGGAAGGUGCUAAUUCCGCAGAGAACACGUUGUGUUUUUUGGAAGAUGA .(((((....(((((..((((.....)))).....)))))((((.(((((((((....---------)))))))..))))))..(((((..((((((((...)))))))))))))))))) ( -38.80) >Bcere.0 2536324 111 + 5224283/0-120 CUUAUCCAGAGAGGUGGAGGGAACGGCCCUAUGAAACCUCAGCAACCCCUAUAUAUAU---------UUAUAGGAAGGUGCUAAUUCCGCAGAGAACACGAUGUGUUUUUUGGAAGAUAA ..((((....(((((..((((.....)))).....)))))((((.((((((((.....---------.))))))..))))))..(((((..((((((((...))))))))))))))))). ( -36.20) >consensus CUUAUCCAGAGAGGUGGAGGGAACGGCCCUAUGAAACCUCAGCAACCCCUAUGUAUAG_________GUACACCAAGGUGCUAAAUCCACAAAGAACACGUUAUGCUU___GGAAGAUAA .(((((....(((((.(((((.....)))))....)))))((((.(((((..(((((((.....))))))))))..))))))...((((.(((((((.....))))))).)))).))))) (-32.46 = -33.02 + 0.56)

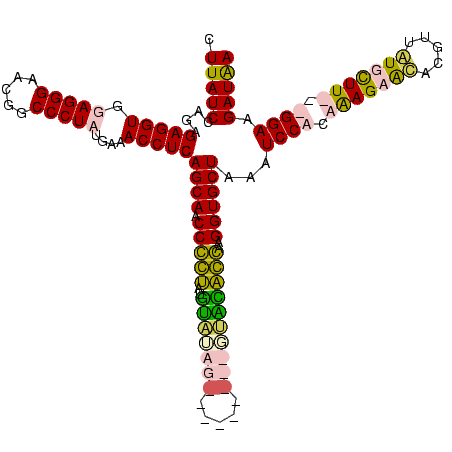
| Location | 1,291,286 – 1,291,398 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.11 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -22.08 |
| Energy contribution | -24.20 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.985002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 1291286 112 + 4222334/0-120 UUAUCUUCC---AAGCAU-----CCGCUUGCUGGAUUUAGCACCUUGGUGCACUGACGUCGUCAGUGCAGACCGGUUGCUGAGGUUUCAAAGGGCCAGUCCCUCCACCUCUCGUGAUAAG (((((.(((---(.(((.-----.....)))))))...((((((..(((((((((((...))))))))..))))).))))(((((.....((((.....))))..)))))....))))). ( -39.40) >Bsubt.0 1257615 116 + 4214630/0-120 UUAUCUUCC---AAGCAUAAAAAACGCUUGCUGGAUUUAGCACCUUGGCGCUUUGCAAAUG-CAAAACAAGCCGGUUGCUGAGGUUUCUAAGGGCCAGUCCCUCCACCUCUCACGAUAAG (((((.(((---(.(((...........)))))))...((((((..(((..(((((....)-))))....))))).))))(((((.....((((.....))))..)))))....))))). ( -33.10) >Banth.0 2459361 111 + 5227293/0-120 UCAUCUUCCAAAAAACACAACGUGUUCUCUGCGGAAUUAGCACCUUCCUAUGC---------AUUUACAUAGGGGUUGCUGAGGUUUCAUAGGGCCGUUCCCUCCACCUCUCUGGAUAAG ..................((((.(((((.((..(..((((((((..((((((.---------.....)))))))).))))))..)..)).)))))))))...((((......)))).... ( -28.70) >Bcere.0 2536324 111 + 5224283/0-120 UUAUCUUCCAAAAAACACAUCGUGUUCUCUGCGGAAUUAGCACCUUCCUAUAA---------AUAUAUAUAGGGGUUGCUGAGGUUUCAUAGGGCCGUUCCCUCCACCUCUCUGGAUAAG (((((........(((((...))))).....((((...((((((..((((((.---------.....)))))))).))))(((((.....((((.....))))..)))))))))))))). ( -28.00) >consensus UUAUCUUCC___AAACACAACGUGCGCUCGCCGGAAUUAGCACCUUCCUACAC_________AAAUACAUACCGGUUGCUGAGGUUUCAAAGGGCCAGUCCCUCCACCUCUCUGGAUAAG (((((.(((...((((((.....))))))...)))...((((((..((((((((((.....)))))))..))))).))))(((((.....((((.....))))..)))))....))))). (-22.08 = -24.20 + 2.12)

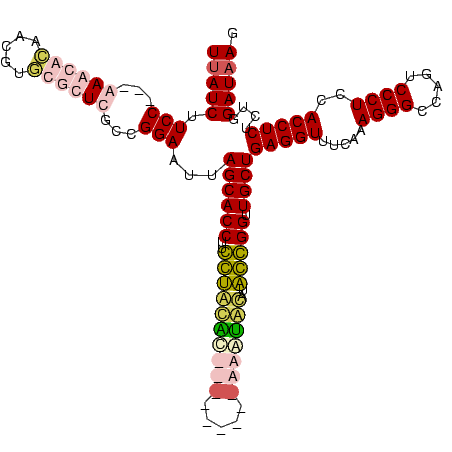
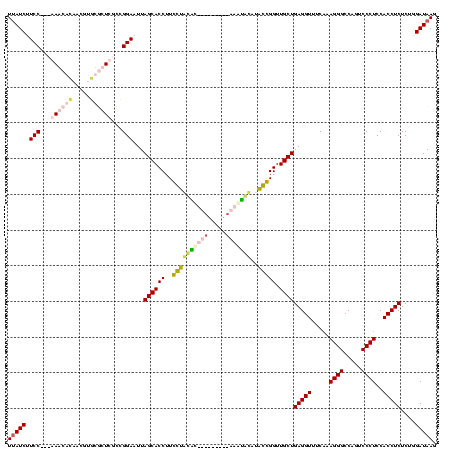
| Location | 1,291,286 – 1,291,392 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 60.71 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -24.65 |
| Energy contribution | -24.21 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.43 |
| Mean z-score | -3.83 |
| Structure conservation index | 0.68 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 1291286 106 + 4222334/40-160 AGCAACCGGUCUGCACUGACGACGUCAGUGCACCAAGGUGCUAAAUCCAGCAAGCGG-----AUGCUU---GGAAGAUAAGAAGA------AGCGAAUAAUCCCCCUUCUUCUUAUGAAG ((((.((....(((((((((...)))))))))....))))))...(((((((.....-----.))).)---)))..(((((((((------((.(........).))))))))))).... ( -39.80) >Bsubt.0 1257615 108 + 4214630/40-160 AGCAACCGGCUUGUUUUG-CAUUUGCAAAGCGCCAAGGUGCUAAAUCCAGCAAGCGUUUUUUAUGCUU---GGAAGAUAAGAAGA------AGCGUUAAA--CCCCUUCUUCUUAUGAAG ((((.(((((..((((((-(....))))))))))..))))))........(((((((.....))))))---)....(((((((((------((.(.....--.).))))))))))).... ( -37.10) >Banth.0 2459361 111 + 5227293/40-160 AGCAACCCCUAUGUAAAU---------GCAUAGGAAGGUGCUAAUUCCGCAGAGAACACGUUGUGUUUUUUGGAAGAUGAGAGGAUUCUUGAACGUGAAAGAAAAUGACCUCUUAUGUAA ((((.(((((((((....---------)))))))..))))))..(((((..((((((((...))))))))))))).((((((((.(((((........))))).....)))))))).... ( -35.00) >Bcere.0 2536324 111 + 5224283/40-160 AGCAACCCCUAUAUAUAU---------UUAUAGGAAGGUGCUAAUUCCGCAGAGAACACGAUGUGUUUUUUGGAAGAUAAGAGGAUUCUUGAACGUGAAAGAAAAUGACCUCUUAUGUAA ((((.((((((((.....---------.))))))..))))))..(((((..((((((((...))))))))))))).((((((((.(((((........))))).....)))))))).... ( -32.40) >consensus AGCAACCCCUAUGUAUAG_________GUACACCAAGGUGCUAAAUCCACAAAGAACACGUUAUGCUU___GGAAGAUAAGAAGA______AACGUGAAAGAAAACGACCUCUUAUGAAA ((((.(((((..(((((((.....))))))))))..))))))...((((.(((((((.....))))))).))))..((((((((........................)))))))).... (-24.65 = -24.21 + -0.44)

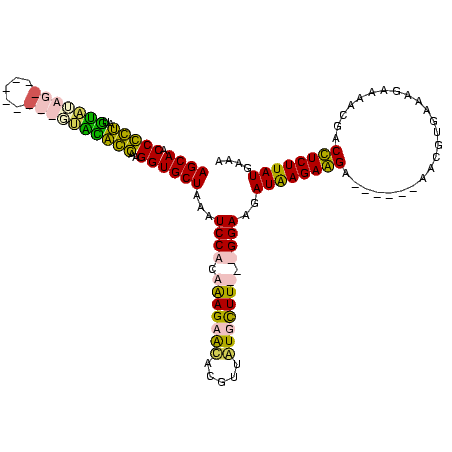
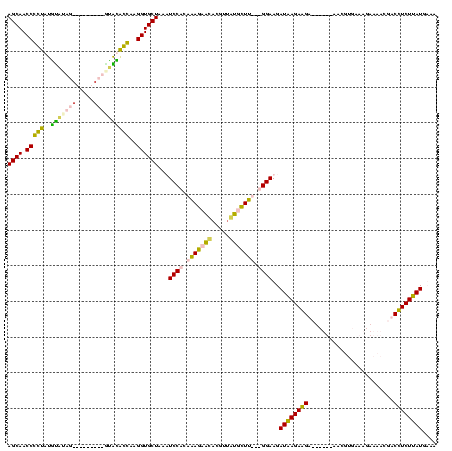
| Location | 1,291,286 – 1,291,392 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 60.71 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -16.16 |
| Energy contribution | -17.79 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.52 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 1291286 106 + 4222334/40-160 CUUCAUAAGAAGAAGGGGGAUUAUUCGCU------UCUUCUUAUCUUCC---AAGCAU-----CCGCUUGCUGGAUUUAGCACCUUGGUGCACUGACGUCGUCAGUGCAGACCGGUUGCU .....((((((((((.(((....))).))------))))))))(((..(---((((..-----..)))))..)))...((((((..(((((((((((...))))))))..))))).)))) ( -40.90) >Bsubt.0 1257615 108 + 4214630/40-160 CUUCAUAAGAAGAAGGGG--UUUAACGCU------UCUUCUUAUCUUCC---AAGCAUAAAAAACGCUUGCUGGAUUUAGCACCUUGGCGCUUUGCAAAUG-CAAAACAAGCCGGUUGCU .....((((((((((.(.--.....).))------))))))))(((..(---((((.........)))))..)))...((((((..(((..(((((....)-))))....))))).)))) ( -35.50) >Banth.0 2459361 111 + 5227293/40-160 UUACAUAAGAGGUCAUUUUCUUUCACGUUCAAGAAUCCUCUCAUCUUCCAAAAAACACAACGUGUUCUCUGCGGAAUUAGCACCUUCCUAUGC---------AUUUACAUAGGGGUUGCU .......(((((.....(((((........))))).)))))....((((....(((((...)))))......))))..((((((..((((((.---------.....)))))))).)))) ( -23.40) >Bcere.0 2536324 111 + 5224283/40-160 UUACAUAAGAGGUCAUUUUCUUUCACGUUCAAGAAUCCUCUUAUCUUCCAAAAAACACAUCGUGUUCUCUGCGGAAUUAGCACCUUCCUAUAA---------AUAUAUAUAGGGGUUGCU ....((((((((.....(((((........))))).)))))))).((((....(((((...)))))......))))..((((((..((((((.---------.....)))))))).)))) ( -25.10) >consensus CUACAUAAGAAGAAAGGGUCUUUCACGCU______UCCUCUUAUCUUCC___AAACACAACGUGCGCUCGCCGGAAUUAGCACCUUCCUACAC_________AAAUACAUACCGGUUGCU ....((((((((........................))))))))..(((...((((((.....))))))...)))...((((((..((((((((((.....)))))))..))))).)))) (-16.16 = -17.79 + 1.63)

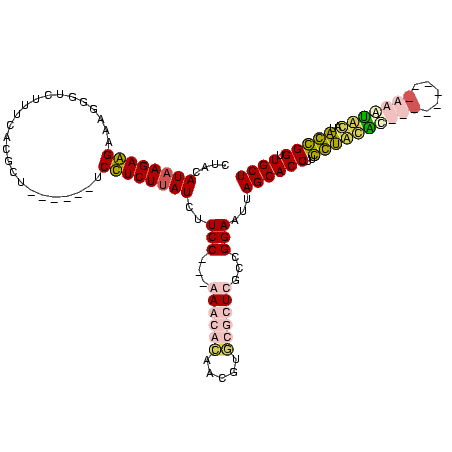
| Location | 1,291,286 – 1,291,392 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.29 |
| Mean single sequence MFE | -35.55 |
| Consensus MFE | -15.40 |
| Energy contribution | -14.46 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.38 |
| Mean z-score | -3.81 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 1291286 106 + 4222334/57-177 GACGACGUCAGUGCACCAAGGUGCUAAAUCCAGCAAGCGG-----AUGCUU---GGAAGAUAAGAAGA------AGCGAAUAAUCCCCCUUCUUCUUAUGAAGAAGGGGUUUUUACUUUU ......(((...((((....))))....(((((((.....-----.))).)---))).)))....(((------((..((.....((((((((((....))))))))))..))..))))) ( -34.80) >Bsubt.0 1257615 108 + 4214630/57-177 G-CAUUUGCAAAGCGCCAAGGUGCUAAAUCCAGCAAGCGUUUUUUAUGCUU---GGAAGAUAAGAAGA------AGCGUUAAA--CCCCUUCUUCUUAUGAAGAAGGGGUUUUUAUUUUG .-.....((..(((((....)))))..(((...(((((((.....))))))---)...))).......------.))...(((--((((((((((....)))))))))))))........ ( -39.00) >Banth.0 2459361 111 + 5227293/57-177 U---------GCAUAGGAAGGUGCUAAUUCCGCAGAGAACACGUUGUGUUUUUUGGAAGAUGAGAGGAUUCUUGAACGUGAAAGAAAAUGACCUCUUAUGUAAGAGGUCAUUUUUUGUUG .---------((((......))))...(((((((((((((((...))))))))))(((..(....)..))).....)).)))(((((((((((((((....))))))))))))))).... ( -35.00) >Bcere.0 2536324 111 + 5224283/57-177 U---------UUAUAGGAAGGUGCUAAUUCCGCAGAGAACACGAUGUGUUUUUUGGAAGAUAAGAGGAUUCUUGAACGUGAAAGAAAAUGACCUCUUAUGUAAGAGGUCAUUUUUUGUUG (---------((((.((((........)))).((((((((((...)))))))))).....((((......))))...)))))(((((((((((((((....))))))))))))))).... ( -33.40) >consensus G_________GUACACCAAGGUGCUAAAUCCACAAAGAACACGUUAUGCUU___GGAAGAUAAGAAGA______AACGUGAAAGAAAACGACCUCUUAUGAAAAAGGGCAUUUUAUGUUG ...............((...........((((.(((((((.....))))))).))))..((((((((........................))))))))........))........... (-15.40 = -14.46 + -0.94)

| Location | 1,291,286 – 1,291,392 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 61.29 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -8.00 |
| Energy contribution | -11.00 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.26 |
| Mean z-score | -4.15 |
| Structure conservation index | 0.28 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 1291286 106 + 4222334/57-177 AAAAGUAAAAACCCCUUCUUCAUAAGAAGAAGGGGGAUUAUUCGCU------UCUUCUUAUCUUCC---AAGCAU-----CCGCUUGCUGGAUUUAGCACCUUGGUGCACUGACGUCGUC ...(((((...((((((((((....))))))))))..)))))(((.------((.....((((..(---((((..-----..)))))..))))...((((....))))...)).).)).. ( -35.10) >Bsubt.0 1257615 108 + 4214630/57-177 CAAAAUAAAAACCCCUUCUUCAUAAGAAGAAGGGG--UUUAACGCU------UCUUCUUAUCUUCC---AAGCAUAAAAAACGCUUGCUGGAUUUAGCACCUUGGCGCUUUGCAAAUG-C ((((....(((((((((((((....))))))))))--)))..((((------....((.((((..(---((((.........)))))..))))..))......)))).))))......-. ( -34.10) >Banth.0 2459361 111 + 5227293/57-177 CAACAAAAAAUGACCUCUUACAUAAGAGGUCAUUUUCUUUCACGUUCAAGAAUCCUCUCAUCUUCCAAAAAACACAACGUGUUCUCUGCGGAAUUAGCACCUUCCUAUGC---------A ......(((((((((((((....)))))))))))))..(((.(((...((((..(.......................)..))))..))))))...(((........)))---------. ( -22.90) >Bcere.0 2536324 111 + 5224283/57-177 CAACAAAAAAUGACCUCUUACAUAAGAGGUCAUUUUCUUUCACGUUCAAGAAUCCUCUUAUCUUCCAAAAAACACAUCGUGUUCUCUGCGGAAUUAGCACCUUCCUAUAA---------A ......(((((((((((((....)))))))))))))..(((.(((..((((....))))...........(((((...)))))....)))))).................---------. ( -21.70) >consensus CAAAAAAAAAACACCUCCUACAUAAGAAGAAAGGGUCUUUCACGCU______UCCUCUUAUCUUCC___AAACACAACGUGCGCUCGCCGGAAUUAGCACCUUCCUACAC_________A ...........((((((((((....))))))))))............................(((...((((((.....))))))...)))............................ ( -8.00 = -11.00 + 3.00)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:25:03 2006