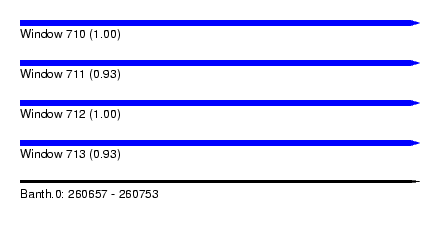
| Sequence ID | Banth.0 |
|---|---|
| Location | 260,657 – 260,753 |
| Length | 96 |
| Max. P | 0.999251 |

| Location | 260,657 – 260,753 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 64.57 |
| Mean single sequence MFE | -20.14 |
| Consensus MFE | -20.54 |
| Energy contribution | -16.86 |
| Covariance contribution | -3.68 |
| Combinations/Pair | 1.90 |
| Mean z-score | -0.57 |
| Structure conservation index | 1.02 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
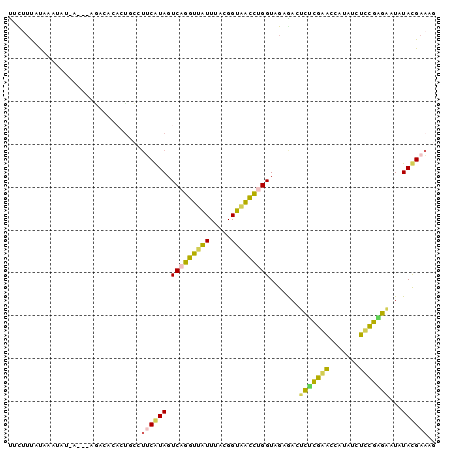
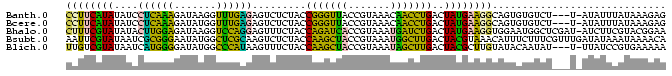
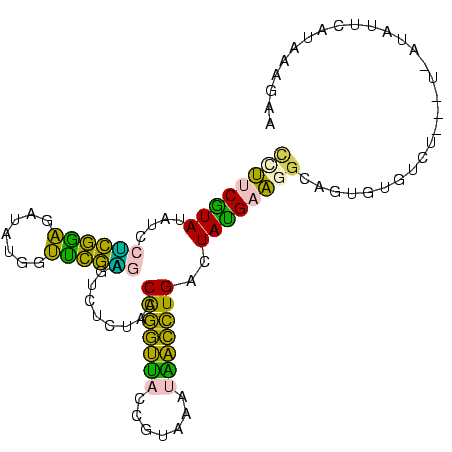
>Banth.0 260657 96 + 5227293 CCUUCAUAUAUCCUCAAAGAUAAGGUUUGAGAGUCUCUACCGGGUUACCGUAAACAACCUGACUAUGAAGGCAGUGUGUCU---U-AUAUUUAUAAAGAG ((((((((....((((((.......)))))).........((((((.........))))))..))))))))..........---.-.............. ( -18.50) >Bcere.0 294525 96 + 5224283 CCUUCAUAUAUCCUCAAAGAUAUGGUUUGAGAGUCUCUACCGGGUUACCGUAAACAACCUGACUAUGAAGGCAGUGUGUCU---U-AUAUUUAUAAAGAG ((((((((....((((((.......)))))).........((((((.........))))))..))))))))..........---.-.............. ( -18.50) >Bhalo.0 648458 99 + 4202352 CUUUCGUAUAUACUUGGAGAUAAGGUCCAGGAGUUUCUACCAGAUCACCGUAAAUGAUCUGACUAUGAAGGUGGAAUGGCUCGAU-AUCUUCGUACGGAA ..(((((((......(((((((.(..(((.....(((((((((((((.......))))))).((....)))))))))))..)..)-))))))))))))). ( -25.90) >Bsubt.0 693791 100 + 4214630 AAUUCGUAUAAUCGCGGGAAUAUGGCUCGCAAGUCUCUACCAAGCUACCGUAAAUGGCUUGACUACGUAAACAUUUCUUUCGUUUGAUAUAAAUAAAACA ....((((.....(((((.......)))))..........(((((((.......)))))))..))))..............((((..........)))). ( -17.10) >Blich.0 693214 96 + 4222334 UUGUCGUAUAAUCAUGGGGAUAUGGCCCAUAAGUUUCUACCAAGCUACCGUAAAUAGCUUGACUACGCUUGUAUACAAUAU---U-UUAUCCGUGAAAAA ...........((((((((((((.....((((((......(((((((.......))))))).....)))))).....))))---)-...))))))).... ( -20.70) >consensus CCUUCGUAUAUCCUCGGAGAUAUGGUUCGAGAGUCUCUACCAGGUUACCGUAAAUAACCUGACUAUGAAGGCAGUGUGUCU___U_AUAUUCAUAAAGAA ((((((((....((((((.......)))))).........(((((((.......)))))))..))))))))............................. (-20.54 = -16.86 + -3.68)
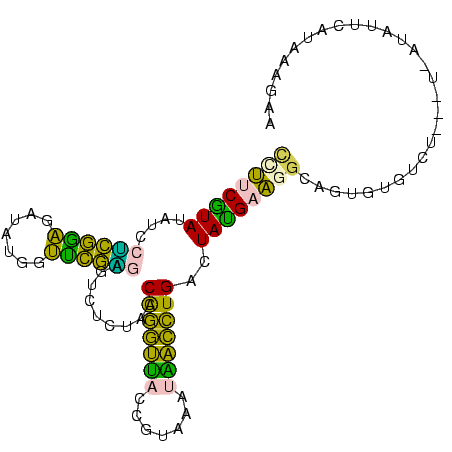

| Location | 260,657 – 260,753 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 64.57 |
| Mean single sequence MFE | -19.22 |
| Consensus MFE | -16.08 |
| Energy contribution | -13.44 |
| Covariance contribution | -2.64 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.45 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
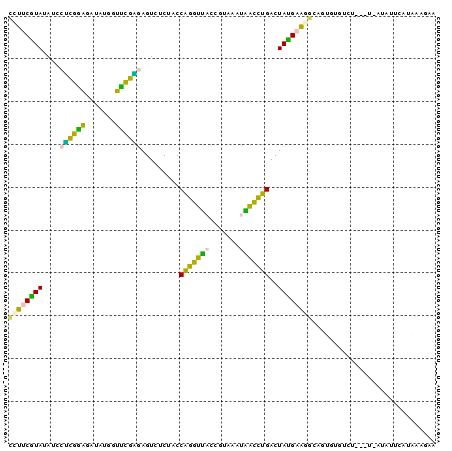
>Banth.0 260657 96 + 5227293 CUCUUUAUAAAUAU-A---AGACACACUGCCUUCAUAGUCAGGUUGUUUACGGUAACCCGGUAGAGACUCUCAAACCUUAUCUUUGAGGAUAUAUGAAGG ..............-.---..........(((((((((((.((((.....(((....))).....))))((((((.......))))))))).)))))))) ( -20.80) >Bcere.0 294525 96 + 5224283 CUCUUUAUAAAUAU-A---AGACACACUGCCUUCAUAGUCAGGUUGUUUACGGUAACCCGGUAGAGACUCUCAAACCAUAUCUUUGAGGAUAUAUGAAGG ..............-.---..........(((((((((((.((((.....(((....))).....))))((((((.......))))))))).)))))))) ( -20.80) >Bhalo.0 648458 99 + 4202352 UUCCGUACGAAGAU-AUCGAGCCAUUCCACCUUCAUAGUCAGAUCAUUUACGGUGAUCUGGUAGAAACUCCUGGACCUUAUCUCCAAGUAUAUACGAAAG ....((((..((((-(.....(((.......(((.((.(((((((((.....)))))))))))))).....)))....)))))....))))...(....) ( -18.90) >Bsubt.0 693791 100 + 4214630 UGUUUUAUUUAUAUCAAACGAAAGAAAUGUUUACGUAGUCAAGCCAUUUACGGUAGCUUGGUAGAGACUUGCGAGCCAUAUUCCCGCGAUUAUACGAAUU ..................(....).........((((.((((((.((.....)).)))))).......(((((...........)))))...)))).... ( -13.90) >Blich.0 693214 96 + 4222334 UUUUUCACGGAUAA-A---AUAUUGUAUACAAGCGUAGUCAAGCUAUUUACGGUAGCUUGGUAGAAACUUAUGGGCCAUAUCCCCAUGAUUAUACGACAA ..............-.---...(((((((.........(((((((((.....))))))))).......(((((((.......))))))).)))))))... ( -21.70) >consensus UUCUUUAUAAAUAU_A___AGACACACUGCCUUCAUAGUCAGGUUAUUUACGGUAACCUGGUAGAGACUCUCGAACCAUAUCUCCGAGAAUAUACGAAAG ...............................((((((.(((((((((.....))))))))).......(((((((.......)))))))...)))))).. (-16.08 = -13.44 + -2.64)

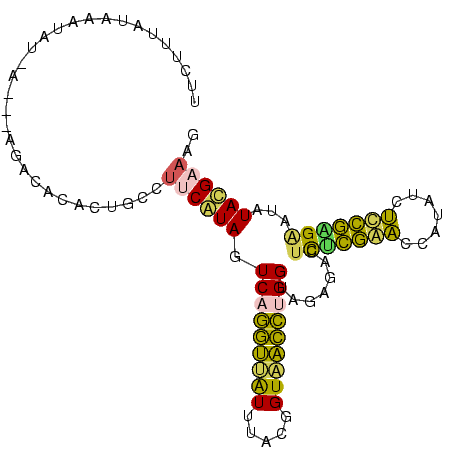
| Location | 260,657 – 260,753 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 64.57 |
| Mean single sequence MFE | -20.14 |
| Consensus MFE | -20.54 |
| Energy contribution | -16.86 |
| Covariance contribution | -3.68 |
| Combinations/Pair | 1.90 |
| Mean z-score | -0.57 |
| Structure conservation index | 1.02 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 260657 96 + 5227293/0-100 CCUUCAUAUAUCCUCAAAGAUAAGGUUUGAGAGUCUCUACCGGGUUACCGUAAACAACCUGACUAUGAAGGCAGUGUGUCU---U-AUAUUUAUAAAGAG ((((((((....((((((.......)))))).........((((((.........))))))..))))))))..........---.-.............. ( -18.50) >Bcere.0 294525 96 + 5224283/0-100 CCUUCAUAUAUCCUCAAAGAUAUGGUUUGAGAGUCUCUACCGGGUUACCGUAAACAACCUGACUAUGAAGGCAGUGUGUCU---U-AUAUUUAUAAAGAG ((((((((....((((((.......)))))).........((((((.........))))))..))))))))..........---.-.............. ( -18.50) >Bhalo.0 648458 99 + 4202352/0-100 CUUUCGUAUAUACUUGGAGAUAAGGUCCAGGAGUUUCUACCAGAUCACCGUAAAUGAUCUGACUAUGAAGGUGGAAUGGCUCGAU-AUCUUCGUACGGAA ..(((((((......(((((((.(..(((.....(((((((((((((.......))))))).((....)))))))))))..)..)-))))))))))))). ( -25.90) >Bsubt.0 693791 100 + 4214630/0-100 AAUUCGUAUAAUCGCGGGAAUAUGGCUCGCAAGUCUCUACCAAGCUACCGUAAAUGGCUUGACUACGUAAACAUUUCUUUCGUUUGAUAUAAAUAAAACA ....((((.....(((((.......)))))..........(((((((.......)))))))..))))..............((((..........)))). ( -17.10) >Blich.0 693214 96 + 4222334/0-100 UUGUCGUAUAAUCAUGGGGAUAUGGCCCAUAAGUUUCUACCAAGCUACCGUAAAUAGCUUGACUACGCUUGUAUACAAUAU---U-UUAUCCGUGAAAAA ...........((((((((((((.....((((((......(((((((.......))))))).....)))))).....))))---)-...))))))).... ( -20.70) >consensus CCUUCGUAUAUCCUCGGAGAUAUGGUUCGAGAGUCUCUACCAGGUUACCGUAAAUAACCUGACUAUGAAGGCAGUGUGUCU___U_AUAUUCAUAAAGAA ((((((((....((((((.......)))))).........(((((((.......)))))))..))))))))............................. (-20.54 = -16.86 + -3.68)
| Location | 260,657 – 260,753 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 64.57 |
| Mean single sequence MFE | -19.22 |
| Consensus MFE | -16.08 |
| Energy contribution | -13.44 |
| Covariance contribution | -2.64 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.45 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 260657 96 + 5227293/0-100 CUCUUUAUAAAUAU-A---AGACACACUGCCUUCAUAGUCAGGUUGUUUACGGUAACCCGGUAGAGACUCUCAAACCUUAUCUUUGAGGAUAUAUGAAGG ..............-.---..........(((((((((((.((((.....(((....))).....))))((((((.......))))))))).)))))))) ( -20.80) >Bcere.0 294525 96 + 5224283/0-100 CUCUUUAUAAAUAU-A---AGACACACUGCCUUCAUAGUCAGGUUGUUUACGGUAACCCGGUAGAGACUCUCAAACCAUAUCUUUGAGGAUAUAUGAAGG ..............-.---..........(((((((((((.((((.....(((....))).....))))((((((.......))))))))).)))))))) ( -20.80) >Bhalo.0 648458 99 + 4202352/0-100 UUCCGUACGAAGAU-AUCGAGCCAUUCCACCUUCAUAGUCAGAUCAUUUACGGUGAUCUGGUAGAAACUCCUGGACCUUAUCUCCAAGUAUAUACGAAAG ....((((..((((-(.....(((.......(((.((.(((((((((.....)))))))))))))).....)))....)))))....))))...(....) ( -18.90) >Bsubt.0 693791 100 + 4214630/0-100 UGUUUUAUUUAUAUCAAACGAAAGAAAUGUUUACGUAGUCAAGCCAUUUACGGUAGCUUGGUAGAGACUUGCGAGCCAUAUUCCCGCGAUUAUACGAAUU ..................(....).........((((.((((((.((.....)).)))))).......(((((...........)))))...)))).... ( -13.90) >Blich.0 693214 96 + 4222334/0-100 UUUUUCACGGAUAA-A---AUAUUGUAUACAAGCGUAGUCAAGCUAUUUACGGUAGCUUGGUAGAAACUUAUGGGCCAUAUCCCCAUGAUUAUACGACAA ..............-.---...(((((((.........(((((((((.....))))))))).......(((((((.......))))))).)))))))... ( -21.70) >consensus UUCUUUAUAAAUAU_A___AGACACACUGCCUUCAUAGUCAGGUUAUUUACGGUAACCUGGUAGAGACUCUCGAACCAUAUCUCCGAGAAUAUACGAAAG ...............................((((((.(((((((((.....))))))))).......(((((((.......)))))))...)))))).. (-16.08 = -13.44 + -2.64)

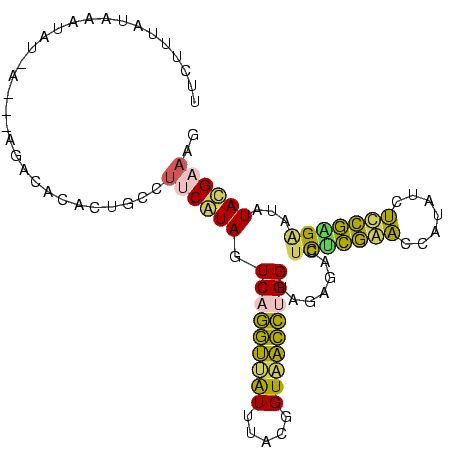
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:24:56 2006